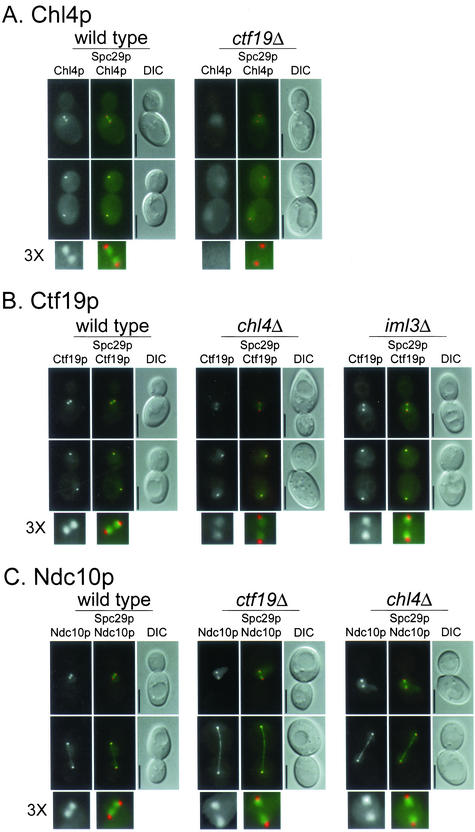
Figure 3.
Interdependence of kinetochore protein localization. (A–E) Indicated proteins were tagged with YFP and imaged as described under MATERIALS AND METHODS. Spc29p-CFP was included in the genetic background as an SPB marker, which allowed determination of the relative location of the kinetochore. In all panels, upper images are cells with short spindles and lower images are cells with long spindles. The left frame has the YFP signal, the middle frame has merged YFP and CFP signals, and the right frame is the corresponding differential interference contrast image. In the color images, YFP is pseudocolored green and CFP is pseudocolored red. A threefold enlargement of the YFP and CFP signals in cells with short spindles, indicated by 3X, is shown at the bottom of each panel. The relevant genetic background (wild-type or kinetochore mutant) is indicated on top of each panel. (F) For colocalization, Chl4p was tagged with YFP and Mif2p was tagged with CFP in a wild-type strain and imaged as in A–E. All strains are homozygous diploids of (A) Chl4p-YFP in wt (YPH1569) and ctf19Δ (YPH1570); (B) Ctf19p-YFP in wt (DHY201), chl4Δ (YPH1571), and iml3Δ (YPH1583); (C) Ndc10p-YFP in wt (YVM1176), ctf19Δ (YPH1572), and chl4Δ (YPH1573); (D) Ctf3p-YFP in wt (DHY202), ctf19Δ (YPH1574), and chl4Δ (YPH1575); (E) Iml3p-YFP in wt (YPH1576), chl4Δ (YPH1577), and ctf19Δ (YPH1578); and (F) Mif2p-YFP (YPH1579) and Chl4p-YFP Mif2p-CFP (YPH1580). All strains (except YPH1580) contain Spc29p-CFP. wt, wild-type. Bar, 5 μm.