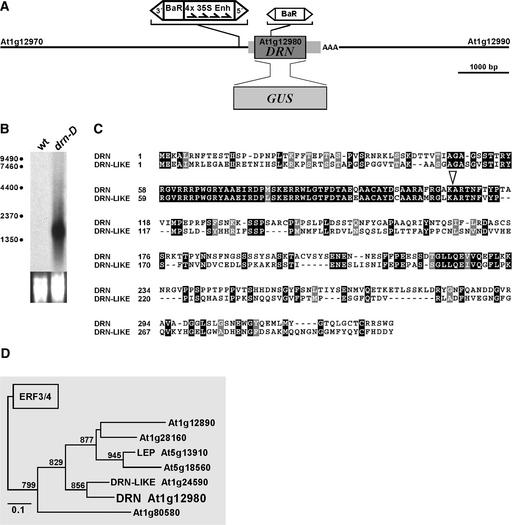
Figure 2.
Structure of the drn-D Allele and Phylogeny.
(A) Position and orientation of the activating en/En transposable element insertion in drn-D relative to the DRN/ESR1 transcription unit and the neighboring At1g12970 and At1g12990 genes. The CaMV35S enhancer elements direct towards the DRN/ESR1 coding region. In the loss-of-function drn-1 allele, a modified dSpm element is inserted into the coding region. The 5′ and 3′ untranslated regions are shown as gray boxes. In the DRN/ESR1-GUS reporter construct, the DRN/ESR1 coding region was replaced by the GUS coding region.
(B) RNA gel blot with 10 μg of total RNA from wild-type (wt) and drn-D seedlings probed for DRN/ESR1 expression. DRN/ESR1 is expressed at high levels in drn-D mutant seedlings. Failure to detect DRN/ESR1 RNA in the wild type is attributable to the very restricted expression domain in the SAM and early primordia anlagen.
(C) DRN/ESR1 protein sequence compared with the sequence of its closest relative, DRN-like (At1g24590). The highly conserved AP2 domain starts at position 56 and ends at residue 116. Within the 68–amino acid AP2 domain, 58 residues are conserved, and a scaffold of Pro (7) and Ser/Thr (11) residues is shared in the C-terminal region. Identical residues are highlighted in black, and isomorphic replacements are highlighted in gray. The open triangle at position 107 indicates the insertion of the dSpm element into the AP2 domain in the drn-1 allele.
(D) DRN/ESR1 is located on a distinct phylogenetic branch of the AP2/ERF transcription factor family. Only one other protein of this family, LEAFY PETIOLE (LEP; van der Graaff et al., 2000), has been analyzed to date.