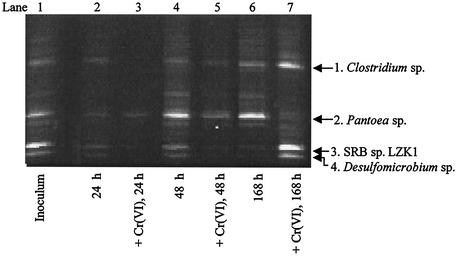
FIG. 2.
Time course PCR-DGGE analysis of the sulfidogenic enrichment consortium MI-S exposed to 0.85 mM Cr(VI). 16S rDNA fragments were analyzed on a denaturing gel (35 to 70% denaturant; Sybr Gold stained). Each lane represents the MI-S community DNA at a specific time point in the absence or presence of 0.85 mM Cr(VI). Because there were considerably fewer cells in the presence of Cr(VI) at 24 h than at other time points, for comparative purposes, 150 ng of total rDNA was loaded into lanes 2 and 3. All other lanes were loaded with 300 ng of total rDNA.