Figure 3.
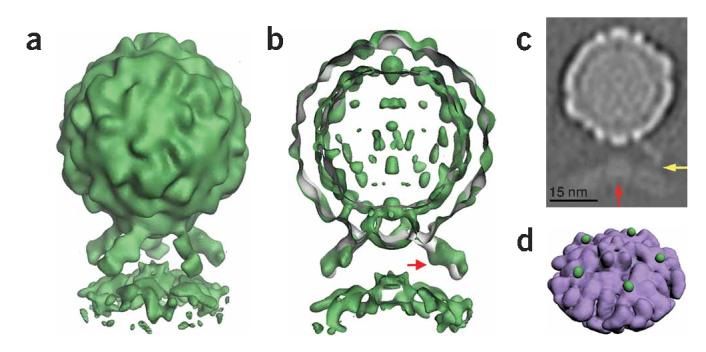
Five-fold symmetric three-dimensional reconstruction of native poliovirus bound to receptor-decorated liposomes. (a) In a surface rendering of the complex (green), the icosahedral features of the virus capsid are maintained, and the density for receptors and membrane are well defined. (b) A cut-away view shows that the three-dimensional reconstruction of the complex (green) superimposes well on electron density (gray) corresponding to the virus-receptor complex in solution. Extra density on the outer leaflet of the membrane near the viral five-fold axis could be due to a perturbation of the membrane. A red arrow indicates a glycosylation site on domain 2 of Pvr. (c) A gray-scale representation of a central slice of the reconstruction that is 27Å thick. This view shows the protrusion of the membrane toward the viral five-fold axis (red arrow) and a weak but continuous density connection between the C-terminal domain of the receptor and the membrane (yellow arrow). Note the clear separation between the inner and outer leaflets of the bilayer, in both the bulged and peripheral areas of the membrane. (d) An unobstructed view of the membrane (purple) shows the crownlike appearance of the apparent distortion. Before removing the receptor density, the small green spheres were located near the predicted C terminus of receptor domain 3. For clarity, d shows lower contour levels than a and b.
