Figure 3.
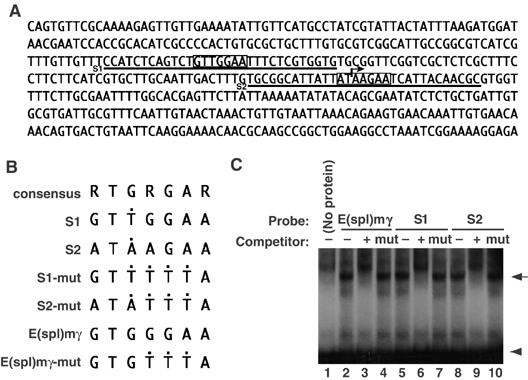
Su(H) binds to the promoter region of chn. (A) The sequence of PR(chn) in the promoter region of chn (Figure 2A). Degenerate Su(H)-binding consensus sites are boxed (S1 and S2, sequences used for EMSA, are underlined). The arrow represents a putative transcriptional initiation site. (B) Core sequences of oligonucleotides used for EMSA. Black dots indicate positions that differ from the Su(H)-binding consensus (5′-RTGRGAR-3′). (C) EMSA using a His fusion of recombinant Su(H). E(spl)mγ is a positive control previously known to bind to Su(H) (lane 1, arrow). Ten-fold excess of unlabeled probe inhibited complex formation (lane 2), but the mutant form of the probe failed to compete (lane 3). S1 and S2 sequences also formed a complex with Su(H) (lanes 4 and 7). Ten-fold excess of unlabeled probes inhibited complex formation (lanes 5 and 8), but competitor containing consensus sequence mutations abrogated the competition (lanes 6 and 9). Arrowhead represents a position of free probe.
