Figure 5.
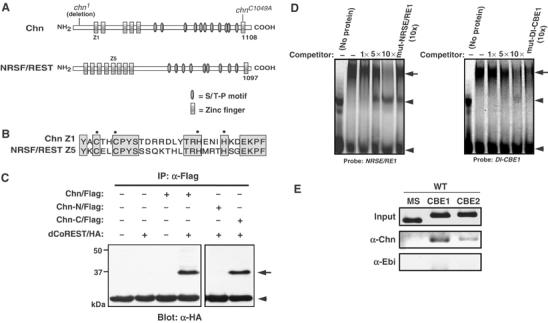
Common properties of Chn and NRSF/REST. (A) Schematic diagram of Chn and human NRSF/REST. Mutation sites of EMS-induced (chn1) and mutagenized (chnC1049A) chn are shown above the Chn diagram. (B) Sequence comparison between the first zinc-finger motif in Chn (Chn Z1) and the fifth zinc-finger motif in human NRSF/REST (NRSF/REST Z5). (C) Immunoprecipitation assay. Chn and dCoREST associate in cultured S2 cells. Chn-N/Flag and Chn-C/Flag contain residues 1–654 and 655–1108, respectively, and are tagged with a single Flag epitope at the C-terminus. dCoREST/HA contains the smallest splice variant of dCoREST, which has EML2 and SANT domains (Dallman et al, 2004) and a single HA epitope at the N-terminus. The arrow indicates the position of dCoREST/HA. The arrowhead indicates the position of IgG. (D) Chn binds to the NRSE/RE1 and Dl-CBE1 sequences. EMSA was performed using NRSE/RE1, a 21-bp recognition sequence of NRSF/REST (Table I), or Dl-CBE1 (Table I) as a probe. A GST fusion of N-terminal zinc-finger domains of Chn formed a specific complex with the probe (arrow). Arrowheads denote a nonspecific shifted band (upper) and free probe (lower). Up to 10-fold molar excess of unlabeled probe was used as competitor. The mutant forms of NRSE/RE1 or Dl-CBE1 failed to compete. (E) ChIP assay of third instar eye imaginal discs. Anti-Chn preferentially precipitated the genomic regions containing CBE1 (Dl CBE1) or CBE2 (Dl CBE2) (Table I), but anti-Ebi did not (WT).
