Figure 3.
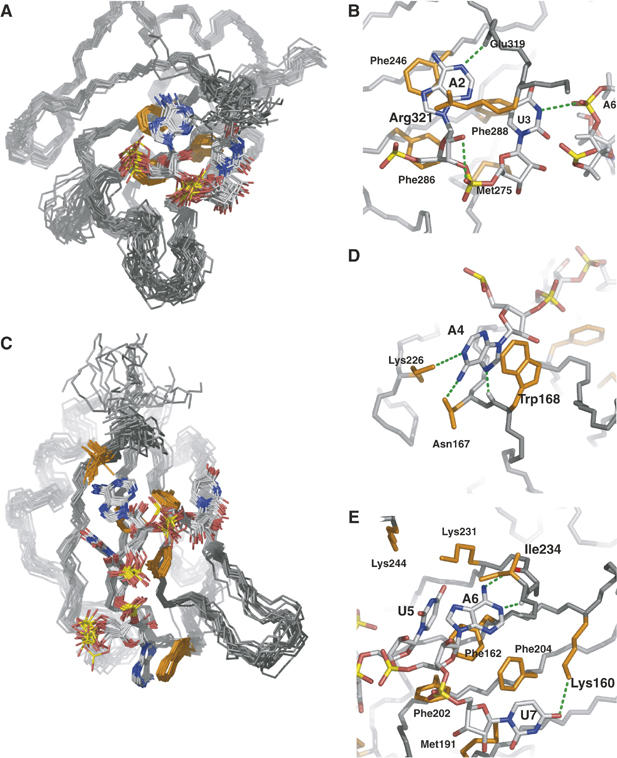
Detailed views of protein–RNA interface of the Hrp1–PEE complex. (A) The C-terminal RBD of Hrp1 recognizes Ade2 and Ura3. (B) Both bases form aromatic interactions with protein residues Phe246 and Phe288. The ribose of Ade2 form van der Waals contacts with Phe286 and Met245, with the last one also contacting the ribose of Ura3. The adenine is specified by a hydrogen bond between the position N1 and the Glu 319 backbone amide whereas the pyrimidine might be specified by a hydrogen bond contact (H3) to Ade6 phosphate group. The ensemble also shows a high population of a ribose-selective hydrogen bond between Ade2 OH2′ and the Ura3 phosphate oxygen. (C) The N-terminal RBD of Hrp1 recognizes four bases from Ade4 to Ura7. (D) One of the novel features of the Hrp1 RNA-binding mode is the well-defined aromatic stacking between Ade4 and Trp168. Adenine selectivity is accomplished by specific hydrogen bonds to positions N1 (Lys226 side chain), NH2(6) (Asn167 side chain) and N7 (Trp168 backbone amide). (E) Another two aromatic stacking interactions between Ade6 and Phe162 and Ura7 and Phe204 dominate the recognition of the 3′-end of the RNA. Ura5 forms van der Waals contacts with Phe162, with two lysine side chains (Lys244 and Lys231) likely involved in the recognition of the carbonyl groups at positions 2 and 4 of the base. Another lysine (Lys160) provides uracil specificity for the seventh base of the RNA by forming a hydrogen bond to the O2 position of this base. Adenine at position five is doubly specified by two hydrogen bonds involving its Watson-Crick face (exocycle NH2 and N1) and protein backbone groups (Ile234 carbonyl and Arg232 amide). Other protein residues (Met191 and Phe216) complete the interactions map by contacting ribose moieties of Ade6 and Ura7.
