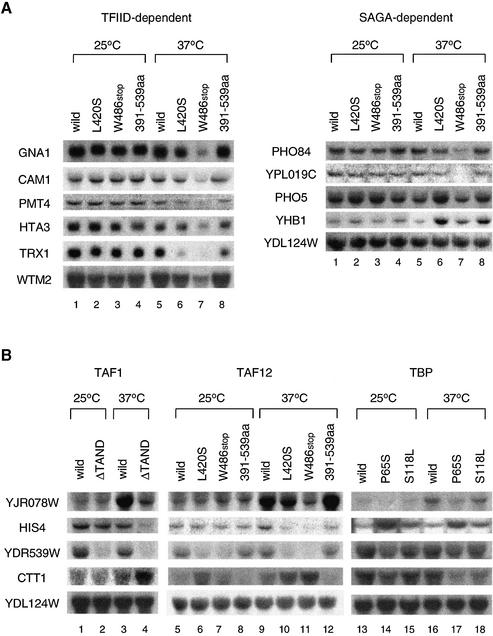
Figure 3.
Transcription analyses of the nsl2/taf12 mutants. (A) The expression of TFIID- and SAGA-dependent genes (35) was compared in three taf12 mutants. Cultures were grown in YPD media to log phase at 25°C; a portion of each culture was shifted to 37°C and incubation continued for 2 h. Total RNA was isolated from wild-type or mutant strains 2 h after a temperature shift to 37°C (lanes 5–8) or continuous incubation at 25°C over the same time period (lanes 1–4). The same amounts of total RNA were blotted onto a nylon membrane and hybridized with the probes indicated. (B) The expression of TAND-dependent genes was compared in the taf1-ΔTAND, nsl2/taf12 and nsl1/spt15 (TBP) mutants. Northern blot analysis was conducted as described in (A) for wild-type or various mutant strains as shown above the lanes using probes of TAND-dependent genes (YJR078W, HIS4, YDR539W and CTT1) and a TAND-independent gene (YDL124W). Although YDL124W was listed as a SAGA-dependent gene by Lee et al. (35), its expression was not changed in the nsl2/taf12 mutants (right panel in A). Thus it serves as an RNA loading control.