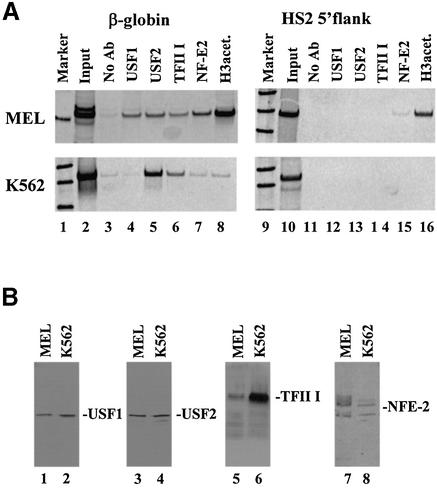
Figure 3.
Characterization of protein–DNA interactions in the human β-globin downstream promoter region in vivo. (A) Chromatin immunoprecipitation (ChIP) experiment analyzing the interaction of proteins with the murine/human β-globin gene or the HS2 5′flanking region in MEL and K562 cells in vivo. Cells were incubated in 1% formaldehyde to crosslink protein–DNA and protein–protein interactions. After sonication, the cells were lysed and the chromatin was precipitated with either no antibody (lanes 3 and 11) or with antibodies specific for USF1 (lanes 4 and 12), USF2 (lanes 5 and 13), TFII-I (lanes 6 and 14), NF-E2 (p45, lanes 7 and 15) or acetylated histone H3 (lanes 8 and 16). DNA was purified from the precipitate and analyzed by PCR for the presence of the murine or human β-globin gene (210 and 321 bp, respectively, lanes 2–8) or the murine or human HS2 5′flank (336 and 565 bp, respectively, lanes 10–16). As positive controls, the input DNA was also analyzed by PCR (lanes 2 and 10). Lanes 1 and 9 show radiolabeled 100 bp markers. (B) Western blot analysis of protein extracts from MEL and K562 cells. Nuclear extracts were prepared from MEL or K562 cells and electrophoresed on denaturing polyacrylamide gels. Proteins were electroblotted to a nitrocellulose membrane, which was then hybridized to antibodies specific for USF1, USF2, TFII-I or NF-E2 (p45) as indicated. Bands were visualized by incubation with horseradish peroxidase-conjugated secondary antibody and autoradiography.