Abstract
The mammalian protein kinase PKR is a critical component of the innate immune response against virus infection. Its cellular actions are mediated by modulating cell signaling and translational regulation. To be enzymatically active, latent PKR needs to be activated by binding to one of its activators, dsRNA or PACT protein. Although the structures of the N-terminal dsRNA-binding domain and the C-terminal kinase domain of PKR have been separately determined, the mode of activation of the enzyme remains unknown. To address this problem, we used biochemical, genetic, and NMR analyses to identify the PACT-binding motif (PBM) located in the kinase domain and demonstrated an intramolecular interaction between PBM and dsRNA-binding domain. This interaction is responsible for keeping PKR in an inactive conformation, because its disruption by point mutations of appropriate residues produced constitutively active PKR. Furthermore, a short decoy peptide, representing PBM, was able to activate PKR by interfering with the intramolecular interaction. These observations suggest a model for PKR activation upon binding of dsRNA or PACT.
Keywords: antiviral mechanism, autoinhibition, NMR, peptide activator, protein kinase
Many cellular functions are regulated by alteration of the phosphorylation status of proteins that mediate them. Consequently, many protein kinases play pivotal roles in the regulation of different aspects of cell growth and metabolism. In this respect, the mammalian protein kinase PKR, which phosphorylates serine and threonine residues of proteins, is the gatekeeper of cellular response to virus infection that is mediated by the products of viral stress-inducible genes. Expression of these genes is induced by cellular exposure to virus infection, IFNs, or dsRNA (1–3), and PKR is required for responding to some of these inducers (4–6). In addition, it is an essential component of the IFN-mediated cellular antiviral response (3, 7). The antiviral effect is achieved by blocking viral protein synthesis as a consequence of PKR-mediated phosphorylation of the α-subunit of the translation initiation factor eIF-2 (8). To evade this effect, many viruses, if not all, encode or induce a variety of inhibitors of PKR, that are proteins or RNAs in nature, indicating the importance of PKR in host defense (3, 9). In addition to this critical role, PKR regulates many other aspects of cellular physiology, such as differentiation, oncogenic transformation, cell growth, and apoptosis (4, 10–13). PKR has also been implicated as a component of many signal transduction pathways used by cytokines, growth factors, dsRNA, and extracellular stresses leading to the activation of transcription factors such as NF-κB, IFN regulatory factor 1, p53, signal transducers and activators of transcription 1 and 3, activating transcription factor (ATF), and AP-1 (14–23). However, for most of these functions of PKR the proximal targets of phosphorylation remain unidentified.
Functions of PKR itself are regulated at two levels. Most cells constitutively express a low level of PKR that is strongly enhanced by cells’ exposure to IFN or virus infection. Another regulation is at the level of enzymatic activation of PKR. PKR remains latent in unstimulated cells, and its activation requires binding of specific activators. It has long been known that viral dsRNA is the primary activator for PKR in virus-infected cells. However, we discovered a few years ago that a human protein, called PACT, could also activate PKR (24), and others have shown that PACT, or its murine counterpart RAX (25), is responsible for cellular PKR activation in response to a variety of extracellular stresses (25–27).
The PKR protein is composed of two distinct domains joined by a linker region: the dsRNA-binding domain (dsRBD) at the N terminus (residues 1–170) containing two dsRNA-binding motifs, dsRBM1 and dsRBM2, and the kinase domain (KD) at the C terminus. Recently, the structure of the PKR KD bound to its substrate, eIF-2α, has been determined (28, 29). These studies demonstrated how phosphorylation of the activation loop of PKR leads to coupling of its distal interfaces that mediates dimerization and substrate binding. The mechanism of dsRNA-mediated PKR activation has received considerable attention over the past several years; however, the molecular basis of PKR activation by PACT remains elusive. Our earlier studies indicated that dsRBM2 may have intramolecular interactions with the KD, thus keeping PKR in a “closed” and inactive conformation; cooperative binding of dsRBM1 and dsRBM2 to dsRNA induces a conformational change of PKR, leading to its activation (30, 31). In contrast, relatively little is known about how PACT activates PKR. PACT contains three independent domains of which the first two resemble dsRBM and the last one (PACTd3), consisting of 66 residues, appears to be a previously unrecognized domain. Mutational analyses have shown that PACTd3 binds weakly to PKR but is necessary and sufficient for activating PKR (32). Domains 1 and 2, on the other hand, may facilitate the process by mediating strong interactions between PACT and PKR (32).
In this study we have used a combined functional and structural approach to determine the mechanism of PACTd3-mediated PKR activation. We show that PACTd3 activates PKR by directly binding to a motif [PACT-binding motif (PBM)] present in the KD. Point mutations within this motif led to constitutive activation of the protein, suggesting that PACTd3 releases PKR from an “inactive” conformation by binding to PBM. We further discovered that the same PBM specifically interacts with dsRBM2, which keeps PKR in a closed conformation. Our results provide mechanistic insights into how PACT binds to PKR in a manner distinct from that of dsRNA and yet the two different bindings lead to the same allosteric changes in PKR and its consequent activation.
Results
Identification of PBM, the PACT Domain 3-Binding Region of PKR.
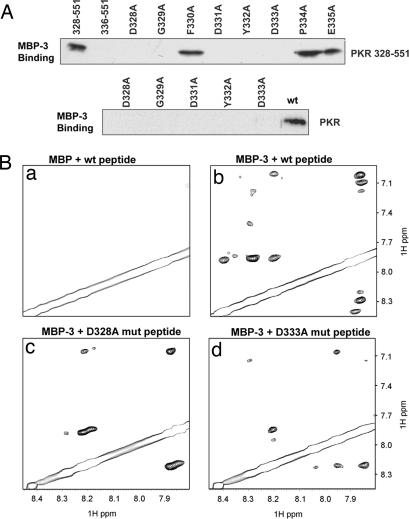
As the first step toward understanding the mechanism of PACTd3-mediated PKR activation we performed a series of experiments to identify the region of PKR to which PACTd3 binds. For this purpose, PACTd3 was expressed in Escherichia coli as a fusion protein with maltose-binding protein (MBP-3) and purified by affinity chromatography. Similarly purified MBP served as the negative control. PKR or its deletion mutants were expressed in human cells as C-terminally FLAG-tagged proteins, purified by affinity chromatography, and challenged for binding to purified MBP-3. MBP-3 and the bound proteins were purified by maltose–agarose chromatography and analyzed by Western blotting using FLAG antibody. As expected, PACTd3 bound to WT PKR, but it also bound to a PKR deletion mutant, missing residues 1–170 (data not shown), to which dsRNA does not bind (33, 34). This result demonstrates that the two activators of PKR, dsRNA and PACTd3, bind to different regions of the protein. Further deletion mapping of PKR revealed that the region between residues 328 and 335 was essential for PACTd3 binding (data not shown). Interestingly, this segment belongs to a nonconservative insert region in the PKR KD (35–37). In the 328–551 PKR mutant, each of the eight residues in this region was individually substituted with alanine, and the mutants were tested for binding to PACTd3; five of the eight alanine substitution mutants failed to bind (Fig. 1A Upper). When the same five point mutations were introduced to full-length PKR, none of the mutants could bind to PACTd3 (Fig. 1A Lower). In all of the above experiments we verified that similar amounts of WT and mutant proteins were expressed and none of the proteins bound to the carrier MBP protein (data not shown).
Fig. 1.
Mapping of the PKR residues needed for PACTd3 binding. (A) Alanine scanning mutagenesis of residues 328–335 of PKR: interaction with PACTd3 measured by coimmunoprecipitation. (Upper) Binding of indicated single-point mutants of FLAG-tagged PKR deletion mutant 328–551 to MBP-3 was measured as described, with PKR WT 328–551 and WT 336–551 serving as positive and negative controls, respectively. (Lower) The mutations in the PKR mutant 328–551 causing a loss of PACTd3 binding activity were duplicated in the context of V5-tagged full-length PKR. Purified WT PKR and the point mutants were assayed for their MBP-3 binding activities; the bound proteins were detected by Western blotting with anti-V5 antibody. (B) Interaction measured by NMR spectroscopy. A selected fingerprint region of 2D NOESY spectra of the PBM peptide (residues 326–337) in the absence and the presence of PACTd3 are presented. (a) Control experiment with the WT peptide in the presence of MBP (no PACTd3). (b) WT peptide and MBP-3. (c) D328A mutant peptide and MBP-3. (d) D333A mutant peptide and MBP-3. Notice substantially reduced transferred NOE peaks in c and d as compared with b, indicating that the mutations reduced the interactions between the peptide and PACTd3.
The above data suggested that PACTd3 might recognize a domain of PKR that contains residues 328–335. To test whether this region can directly bind to PACTd3 we used NMR spectroscopic analysis. The 2D 1H NOESY spectra of the peptide containing residues 326–337 of PKR were recorded in the presence of MBP or MBP-3 (Fig. 1B a and b). The NOESY experiments provide structural information of proteins in the form of nuclear Overhauser effects (NOEs) that reflect 1H–1H contacts within 5 Å. Peptides such as PKR 326–337 usually exhibit few NOEs because of their small sizes. However, in the presence of a large binding partner such as MBP-3, chemical exchange may occur between PKR 326–337 and MBP-3, yielding transferred NOEs on the peptides (for a detailed review on transferred NOEs, see ref. 38). As shown in Fig. 1Bb, MBP-3 (≈50 kDa) indeed caused substantial transferred NOEs to the small peptide as compared with the MBP alone (Fig. 1Ba), indicating that the peptide is involved in interacting with PACTd3. Note that MBP fused to PACTd3 artificially increased the size of the bound peptide/PACTd3 complex, making it feasible to perform high-sensitivity transferred NOE experiments (38). The above experiment led us to define PKR 326–337 as the PACTd3-binding motif (PBM). As expected, two peptides carrying two mutations in this region, D328A and D333A, which, by biochemical assays, were shown to cause a loss of MBP-3 binding (Fig. 1A), had much less interaction with MBP-3 as determined by NMR spectroscopy (Fig. 1B c and d).
Interaction of PBM with dsRBM2.
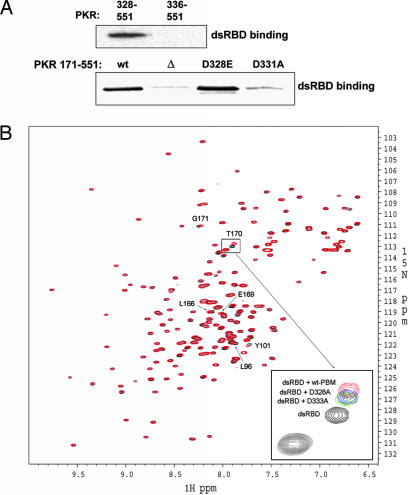
Identification of PBM as the region of the PKR KD with which PACTd3 interacts suggested that the same region might be involved in a conformational regulation of PKR activation. Because it was previously shown that dsRBM2 binds to the C-terminal KD (31), we speculated that PBM might be involved in an intramolecular interaction with dsRBM2, keeping PKR in a closed inactive conformation. To test this hypothesis we performed two experiments shown in Fig. 2. First, the PKR mutant 328–551, but not the mutant 336–551 lacking PBM, interacted with dsRBD, which contains both dsRBM1 and dsRBM2, as demonstrated by their coimmunoprecipitation (Fig. 2A). The same was true for the KD (residues 171–551). It bound dsRBD strongly, but a mutant missing residues 328–335 (Δ) did not (Fig. 2A). A specific point mutation (D331A) that eliminated the interaction of this region with PACTd3 (Fig. 1) also eliminated its interaction with dsRBD. Another mutation (D328E) did not affect the interaction (see also Fig. 3C). More specifically, titration of PBM into 15N-labeled dsRBD induced small but definitive spectral changes of a set of residues (L96, Y101, L104, I105, R107, K112, Y162, L166, E169, T170, and G171) (Fig. 2B). These residues are located on a positively charged surface of dsRBM2 (30, 31), which partially overlaps with the putative dsRNA-binding site (30, 31), indicating specific interactions between dsRBM2 and PBM. In this assay two mutations in the PBM, D328A and D333A, which had reduced its interaction with PACTd3 (Fig. 1), reduced its interaction with dsRBD as well (see Fig. 2B Inset).
Fig. 2.
Interaction of the PBM with dsRBD. (A Upper) The PBM (residues 328–335 of PKR) is needed for binding to dsRBD. V5-tagged dsRBD of PKR was expressed in cells and immunoprecipitated by using anti-V5-agarose. After changing to low-salt binding buffer, purified PKR mutants, 328–551-FLAG and 336–551-FLAG, were added to the beads, and the beads were washed with the binding buffer. The proteins associating with the dsRBD were analyzed by Western blotting with anti-FLAG antibody. (A Lower) Interaction of the KD with dsRBD. The KD including the linker region (residues 171–551) with a FLAG tag was expressed, purified, and tested for binding to dsRBD as above. wt, the KD; Δ, 328–335 deleted from the KD; D328E, that mutation in the KD; D331A, that mutation in the KD. (B) Interaction of the PBM peptide (residues 326–337 of PKR) with dsRBD: NMR spectroscopy. 1H-15N correlation spectra of the dsRBD of PKR in the absence and the presence of the WT and its mutants (D328A and D333A) are presented. 2D 1H-15N heteronuclear single quantum correlation experiments of 0.2 mM 15N-labeled dsRBD in the absence (black) and the presence (red) of 1 mM peptide and its mutants were performed. A cluster of residues in the dsRBD of PKR is perturbed upon addition of the WT peptide (see labels for some of the most perturbed ones), indicating specific interaction between the dsRBD and the peptide. In contrast, the mutant peptides exhibited significantly reduced binding to dsRBD, as shown by the reduced chemical shift changes (see the expanded Inset for T170). T170 was the most perturbed residue when WT peptide was used, but its perturbation was much less with the mutant peptides (see D328A in blue and D333A in green).
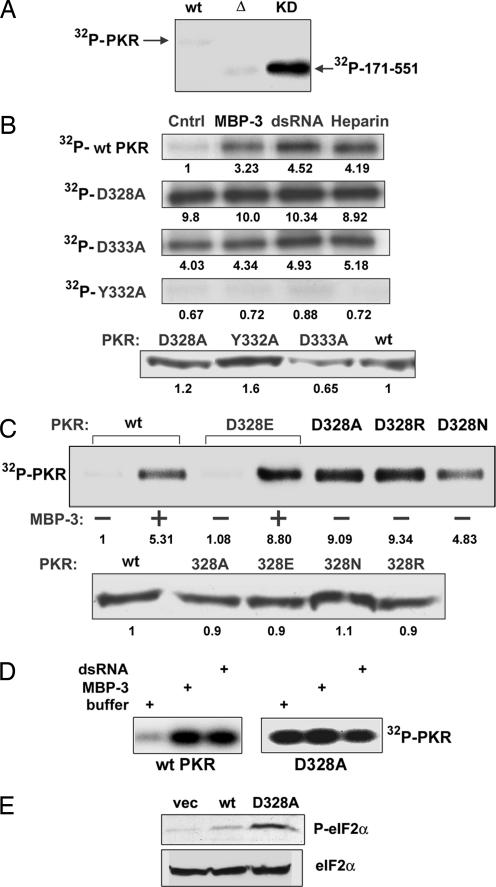
Fig. 3.
Enzymatic activities of PKR mutants as measured by 32P labeling of autophosphorylated proteins. (A) Constitutive activity of the KD. Activities of the isolated KD (residues 171–551) and its mutant were measured in vitro. WT, WT PKR; Δ, the KD missing residues 328–335; KD, the WT KD. (B) Constitutive activation of PKR by mutations of D328 and D333. (Upper) In vitro kinase assays of WT PKR, D328A PKR, D333A PKR, and Y332A PKR were performed in the presence or the absence of MBP-3 (100 nM), dsRNA (200 ng/ml), or heparin (10 units/ml). PKR autophosphorylation was detected by autoradiography. Cntrl, activity buffer. The number at the bottom of each band indicates its relative intensity as measured by PhosphorImager analysis; the intensity of the WT PKR Cntrl was set at 1.0. (Lower) Western blot for PKR and its mutants. The numbers at the bottom are densitometric quantifications of the intensities of the bands. (C) Essential role of the negative charge of D328 in PKR activation. D328 was mutated to alanine, glutamic acid, arginine, or asparagine, and the resultant PKR mutants were examined as in B for their kinase activities (Upper) and protein contents (Lower). (D) Activity of PKR expressed in bacteria. Kinase activities of purified PKR expressed in E. coli were measured in the presence or the absence of the activators. (Left) WT PKR (100 nM). (Right) D328A mutant (100 nM). Where indicated, 100 nM MBP-3 or 200 ng/ml dsRNA was used. (E) Constitutive activity of a PKR mutant in human cells. In vivo action of a constitutively active PKR mutant was tested by expressing it in human cells and measuring the eIF-2α phosphorylation status. (Upper) Western blot with a phospho-eIF-2α-specific antibody. (Lower) Total eIF-2α levels.
Generation of Constitutively Active Mutants of PKR by Disrupting PBM–dsRBM2 Interaction.
If the observed dsRBD/PBM interaction was operational intramolecularly in the PKR protein, it might be instrumental in keeping the protein in the inactive conformation. In that case, one can expect that removal of the dsRBD will make the KD constitutively active. This expectation was experimentally verified by measuring its kinase activity (Fig. 3A). Although, unlike full-length PKR, WT KD was constitutively active, a mutant KD (Δ) missing residues 328–335 was not, indicating that this region, in addition to interacting with dsRBD, may be required for maintaining the structural integrity of the KD. More subtle changes that disrupt dsRBD/PBM interactions should also activate PKR; mutations of appropriate residues in the PBM should convert the protein to a constitutively active form. Indeed, this was the case for the residues D328 and D333. When either of these residues was substituted with alanine, the mutant PKR was fully active as a protein kinase, without an activator, as revealed by its efficient autophosphorylation (Fig. 3B Upper). None of the three known activators of PKR, PACTd3, dsRNA, or heparin, could activate the protein further. In contrast, three other mutants (G329A, D331A, and Y332A) were totally inactive even in the presence of the activators (Fig. 3B and data not shown). The latter mutations might have perturbed the overall structure of the protein in a way that its enzymatic activity was destroyed. In any event, the gain-of-function mutations D328A and D333A were mechanistically more illuminating. Detailed analyses of D328 substitution demonstrated that an acidic residue was required in that position to keep the protein in an inactive conformation because substitution of D328 with glutamic acid did not affect the protein’s properties, whereas substitution of the same residue with alanine, arginine, or asparagine led to constitutive activation of the protein (Fig. 3C). Note that KD containing the D328E mutation, but not the D328A mutation, bound to dsRBD efficiently (see Fig. 2A). The autophosphorylation of WT PKR and its mutants was quantified by phosphorimager analysis of the gels (see the number below each lane in Fig. 3 B and C). WT PKR was activated three to five times by the different activators. The constitutively active mutants were even more active without any activator. The amount of PKR present in each reaction was quantified by Western blotting and found to be similar (Fig. 3 B and C Lower).
Additional experiments were carried out to confirm our hypothesis that the PBM–dsRBD2 interaction keeps PKR in the inactive conformation. WT PKR and the D328A mutant were expressed in E. coli, purified, and tested for enzymatic activities (Fig. 3D). As expected, the WT protein was virtually inactive, but it could be activated strongly by either MBP-3 or dsRNA. In contrast, the D328A mutant PKR was highly active even without any activator. PKR purified from bacteria could be quantified reliably, and we used 100 nM PKR and 100 nM MBP-3 in these reactions. Finally, we tested the activity of this mutant in human cells. Its expression caused pronounced phosphorylation of eIF-2α (Fig. 3E), indicating that the mutant PKR, unlike WT PKR, was constitutively active in human cells.
Activation of PKR by PBM Decoy Peptide.
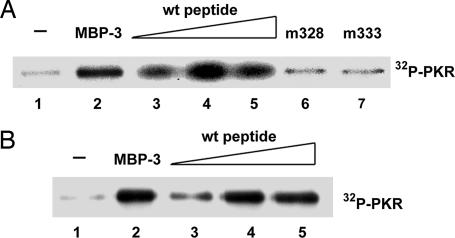
Because a peptide constituting a part of PBM could interact, albeit weakly, with dsRBD (Fig. 2B), we wondered whether this decoy peptide could activate PKR by competing with PBM for its intramolecular interaction with dsRBD. Two sources of purified PKR were used for these experiments, PKR expressed in human cells (Fig. 4A) or in E. coli (Fig. 4B). PKR from both sources was enzymatically inactive as such but could be activated efficiently by MBP-3. As the potential peptide activator, we used a synthetic peptide whose sequence corresponded to residues 326–337 of PKR, the region where PBM is located. This peptide had been shown to interact with dsRBD (see Fig. 2B). As anticipated, the decoy peptide could activate PKR in a dose-dependent fashion. In contrast to the WT peptide, two mutant peptides that interact with dsRBD poorly (see Fig. 2B) failed to activate PKR (Fig. 4A). These results demonstrated that an exogenous decoy peptide could compete with PBM for interaction with dsRBM2; the consequent disruption of the intramolecular interaction between PBM and dsRBM2 caused activation of PKR. Because the mutant peptides could not bind to dsRBM2 efficiently (Fig. 2B), they could not compete out PBM, and hence PKR remained in the inactive conformation. Our results presented here demonstrate that PBM constitutes the intramolecular inhibitory domain, which, by interacting with dsRBD, keeps PKR locked in an inactive conformation.
Fig. 4.
Activation of PKR by the PBM decoy peptide. (A) Activation of PKR purified from human cells. PKR activation assays were done in the presence or the absence of the peptides used in Fig. 2B. Lane 1, no addition; lane 2, 100 nM MBP-3; lane 3, 100 μM WT peptide; lane 4, 500 μM WT peptide; lane 5, 1 mM WT peptide; lane 6, 1 mM D328A mutant peptide; lane 7, 1 mM D333A mutant peptide. (B) Activation of PKR purified from E. coli. Lanes 1–5 were the same as in A except for the source of PKR. PKR at 100 nM was used in all reactions.
Discussion
A variety of genetic, biochemical, and structural evidences has indicated that activation of PKR results from a conformational change of the protein that leads to its dimerization, ATP binding, and autophosphorylation (4, 39–43). These studies also suggested that the latent inactive conformation was maintained by an intramolecular interaction between two regions of the protein (42–44), and our earlier studies indicated that these two regions might be located in the dsRBM2 and the KD, respectively (31). The results presented here confirm that hypothesis, delineate the region in the KD responsible for this interaction, identify specific residues as major contributors to it, and reveal two distinct, but complementary, mechanisms used by dsRNA and PACTd3 to disrupt this interaction and activate PKR.
Our results demonstrate that, unlike dsRNA, PACTd3 does not bind to the dsRBD of PKR; instead it binds to the insert region of the KD. Thus, the two activators bind to the opposite partners of the intramolecular interaction, but both cause its disruption (Fig. 5). Fine mapping of the region required for PACTd3 interaction, PBM, identified five residues within this motif, three aspartic acid, one glycine, and one tyrosine, to be absolutely necessary for the interaction. These residues are well conserved in PKR proteins of other mammals but not in other eIF-2 kinases, indicating that PACTd3 interaction is PKR-specific (37). Surprisingly, PBM also mediated the intramolecular interaction of the KD with dsRBD. As predicted from our earlier study, the interaction was with residues solely in dsRBM2, not dsRBM1. As expected from our model, deletion of the dsRBD produced a constitutively active kinase (Fig. 3A). Similarly, it has been reported that deletion of PKR 159–184 that partly contains the PBM-perturbed region of dsRBD leads to constitutive activation of PKR (44), indicating that the deletion might have disrupted the interaction between dsRBM2 and PBM and led to an open/active conformation of PKR. Although PBM, the region in the KD that interacts with dsRBM2, may extend further toward the C terminus, our evidence clearly shows that residues 328–335 are its major components. A peptide containing these residues interacted with dsRBM2 and with PACTd3; it also functionally blocked the inhibitory action of dsRBM2. The affinity of PBM for PACTd3 must be higher than that for dsRBM2, because when PKR, MBP-3, and the PBM peptide were present together PKR remained inactive (data not shown). In contrast, MBP-3 or the PBM peptide could activate PKR when added alone (Fig. 4). This affinity difference is probably operative for PBM present as a part of PKR as well, thus allowing PACTd3 to pull apart PBM from dsRBM2 and activate the enzyme.
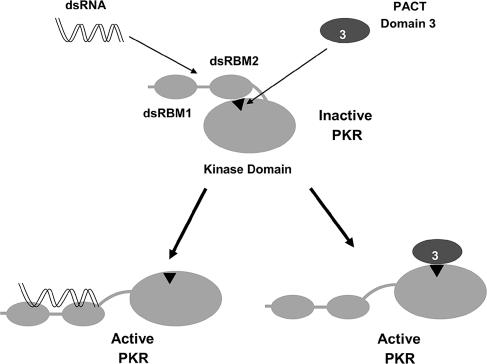
Fig. 5.
Model for PKR activation. The intramolecular inhibitory interaction between the PBM (residues 328–335 of PKR, represented as Δ) in the KD and dsRBM2 maintains PKR in an inactive conformation. Binding of dsRNA to dsRBM1 and dsRBM2 or binding of PACT domain 3 to PBM disrupts the intramolecular interaction in latent PKR and induces similar conformational changes leading to its activation.
Our studies demonstrate that, among the five residues in PBM that are essential for PACTd3 interaction, the acidic residues D328 and D333 are critical; their mutations to neutral or basic residues disrupted the interaction of PBM with dsRBD. The putative roles of the other three essential residues are not clear because their individual mutations or a deletion of the region (328–335) inactivated the protein completely. KDs carrying these mutations did not interact with PACTd3 or dsRBD, and they were enzymatically inactive. Moreover, PKR carrying point mutations in any of those three residues could not be activated, indicating that these residues may be essential for proper folding and/or maintenance of the overall structure of the protein.
The recent structural studies (28, 29) elegantly elucidated the mechanism by which eIF-2α docks on PKR. This process appears to be dynamic and may contribute to the organization of the active site; however, substrate binding is not required for kinase activation. Unfortunately, these investigations used a truncated PKR missing the dsRBD. Thus, they did not address the issue of how PKR gets activated (45). Moreover, a long linker region in the KD was deleted from the analyzed PKR mutant; this missing region contains the PBM, as defined in this article. Thus, the two critical parts of the protein (PBM and dsRBM2) that are directly responsible for keeping PKR locked in the inactive conformation were missing from the PKR mutant analyzed by Dar et al. (28). This study fills an important gap in knowledge by demonstrating an interaction between the PBM and specific residues of dsRBM2 and, more importantly, by establishing that the newly discovered intramolecular interaction is pivotal for keeping PKR in the inactive conformation.
Materials and Methods
Cell Lines and Reagents.
HEK293T and HT1080 cells were cultured in high-glucose DMEM supplemented with 10% (vol/vol) FBS, 2 mM l-glutamine, and antibiotics. Cells were transfected with FuGENE 6 (Roche Diagnostics) or CaPO4 (46). FLAG peptide, anti-FLAG monoclonal M2 antibody, M2-agarose, and protein A-agarose were obtained from Sigma, and V5 peptide, anti-V5 monoclonal antibody, and anti-V5-agarose were from Invitrogen. Antibodies for phospho-eIF-2α and eIF-2α were obtained from Cell Signaling Technology (Beverly, MA).
Expression and Purification of PKR and Its Mutants.
The generation of FLAG- and V5-tagged PKR constructs [pcDNA3-PKR(K296R)-FLAG and pV5-PKR] has been described previously (32, 47, 48). PCR was used to generate FLAG-tagged PKR mutants with a succession of N-terminal deletions that were ligated in-frame into pcDNA3 vector. FLAG-tagged PKR 328–551 mutants with single-point amino acid substitutions were generated by PCR using primers containing desired mutations. The V5-tagged PKR single-point mutants were constructed by overlap extension PCR as previously described (32). Briefly, two separate PCRs were performed to amplify two overlapping halves of the coding region of PKR using four primers. The point mutation was introduced by the middle two primers. The final PCR products were cloned into vector Vet (48). All of the resulting DNA clones were sequenced to verify the desired mutations. PKR or its mutants were expressed in human cells and purified as described in ref. 32. Alternatively, they were expressed in E. coli as hexahistidine-tagged protein and purified by Ni-agarose affinity chromatography using methods described for PACT purification (32).
Assay for Binding of PACT Domain 3 to PKR.
Bacterial expression and purification of MBP and MBP-3 have been described (32). As judged from gel-filtration profile, MBP alone elutes as monomers, whereas MBP-3 elutes as dimers. PKR(K296R)-FLAG and its derivatives were purified by M2-agarose purification as described (47). Bound PKR-FLAG proteins were eluted off the beads with 0.2 mg/ml FLAG peptide (Sigma) in Tris buffer (10 mM Tris·HCl, pH 7.5/10% glycerol). V5-tagged PKR proteins were expressed from transfected HEK293T cells and purified by using methods described for PKR-FLAG purification (47). V5-PKR protein was eluted by using 0.4 mg/ml V5 peptide. For binding assays, 0.5 μg of purified MBP or MBP-3 was incubated with amylose resin (New England Biolabs) in amylose-binding buffer (20 mM Tris·HCl, pH 7.5/10 mM 2-mercaptoethanol/1 mM EDTA/10% glycerol) at 4°C for 1 h. After washing, purified PKR-FLAG or V5-PKR proteins were added to the resin-bound MBP or MBP-3 in amylose-binding buffer and incubated for 1 h at 4°C. The beads were washed six times with amylose-binding buffer containing 1% Triton X-100. Proteins bound to the beads were separated by SDS/PAGE followed by Western blot analyses with anti-FLAG or anti-V5 antibody for detecting PKR-FLAG or V5-PKR.
NMR Experiments.
Uniformly 15N-labeled dsRBD sample was prepared, and the heteronuclear NMR experiments were performed as previously described (49, 50). DsRBD elutes from gel-filtration column as monomers, which is consistent with its overall correlation time (49). The dsRBD sample was prepared in argon-purged H2O solution (7% 2H2O/100 mM NaCl/20 mM sodium phosphate/1 mM DTT, pH 6.5) in a 250-μl microcell NMR tube (Shigemi, Allison Park, PA) at a concentration of ≈0.2 mM. WT PBM peptide and its mutants, D328A and D333A, were synthesized by Lerner Research Institute Biotechnology Core and purified by HPLC. The purified peptides were verified by mass spectroscopy and were then dissolved in the same buffer (1 mM stocks) as dsRBD with the pH adjusted to pH 6.5. Standard heteronuclear single quantum correlation experiments of 0.2 mM 15N-labeled dsRBD in the absence and presence of 1 mM PKR peptide (residues 326–337) and its mutants were performed at 25°C and pH 6.5, using a Bruker ICE 600 MHz spectrometer equipped with a cryogenic triple-resonance probe head and a shielded z-gradient unit. To examine the interaction of the PKR peptide and its mutants with PACTd3, 2D 1H NOESY experiments were performed on 1 mM PKR peptide or its mutants in the absence and presence of 0.1 mM MBP or 0.1 mM MBP-3 at 25°C, 20 mM sodium phosphate. The experiments were run on a Bruker ICE600 equipped with a cryogenic probe. Each experiment took 4.5 h, with a mixing time of 400 ms, and the data points were 1,024 and 256 in F2 and F1 dimensions, respectively.
dsRBD Coimmunoprecipitation Assays.
HEK293T cells were transfected with V5-tagged dsRBD (residues 1–170) of PKR and lysed in TNEN buffer. Extracts containing 1 mg of protein were immunoprecipitated overnight at 4°C with anti-V5-agarose in TNEN buffer. Beads were then washed three times with TNEN buffer and twice with binding buffer (20 mM Tris·HCl, pH 7.5/0.5% Triton X-100/10 mM 2-mercaptoethanol/1 mM EDTA/10% glycerol). After washing, V5-dsRBD associated with the beads was incubated for 2 h at 4°C with FLAG-tagged PKR constructs that were purified as described above. Beads were then washed five times with the binding buffer, and the immunocomplexes were analyzed by Western blotting with antibodies against FLAG and V5 epitopes.
In Vitro Kinase Assay.
The PKR activation assays were performed as described previously (32) using ectopically expressed V5-tagged PKR. HEK293T cells transfected with 10 μg of pV5-PKR by CaPO4 transfection were collected and lysed in TNEN buffer (20 mM Tris/100 mM NaCl/1 mM EDTA/0.5% Nonidet P-40/100 units/ml aprotinin/0.1 M PMSF/100 μM vanadate/1 mM NaF, pH 8.0). Cell lysate containing 100 μg of total protein was mixed with 0.5 μg of anti-V5 antibody in TNEN buffer and rotated at 4°C for overnight. Protein A-agarose was then added for an additional hour followed by washing four times with 500 μl of TNEN buffer and twice with activity buffer (20 mM Tris, pH 7.5/50 mM KCl/2 mM MgCl2/2 mM MnCl2/200 units/ml aprotinin/0.1 mM PMSF/5% glycerol). A kinase assay was performed in activity buffer containing immobilized PKR, an activator (dsRNA, MBP-3, or heparin), and 1 μCi (1 Ci = 37 GBq) of [γ-32P]ATP at 30°C for 30 min. To test the effect of the synthesized WT or mutant peptides (residues 326–337 of PKR) on PKR activation, MBP-3 was replaced by the peptide. Autophosphorylated PKR was analyzed by SDS/PAGE and visualized by autoradiography. The extent of phosphorylation was quantified by phosphorimager. Activity assays for PKR expressed in E. coli were done with purified proteins in solution.
eIF-2 Phosphorylation Assay.
HEK293T cells were transfected with expression vectors of PKR or its mutant. After 48 h cell extracts were made and Western blotted with antibodies for phospho-eIF-2α and eIF-2α.
Acknowledgments
We thank Saurav Misra and Sujay Ithychanda for helpful discussions and Srabani Pal for technical assistance. This study was supported by National Institutes of Health Grants CA62220 and CA68782 (to G.C.S.) and HL58758 and HL073311 (to J.Q.).
Abbreviations
- dsRBD
dsRNA-binding domain
- PBM
PACT-binding motif
- KD
kinase domain
- MBP
maltose-binding protein
- NOE
nuclear Overhauser effect.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. Annu. Rev. Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Sarkar S. N., Peters K. L., Elco C. P., Sakamoto S., Pal S., Sen G. C. Nat. Struct. Mol. Biol. 2004;11:1060–1067. doi: 10.1038/nsmb847. [DOI] [PubMed] [Google Scholar]
- 3.Sarkar S. N., Sen G. C. Pharmacol. Ther. 2004;103:245–259. doi: 10.1016/j.pharmthera.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 4.Williams B. R. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 5.Diebold S. S., Montoya M., Unger H., Alexopoulou L., Roy P., Haswell L. E., Al-Shamkhani A., Flavell R., Borrow P., Reis y Sousa C. Nature. 2003;424:324–328. doi: 10.1038/nature01783. [DOI] [PubMed] [Google Scholar]
- 6.Horng T., Barton G. M., Medzhitov R. Nat. Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 7.Clemens M. J., Elia A. J. Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- 8.Samuel C. E. J. Biol. Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 9.Gale M., Jr, Katze M. G. Pharmacol. Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman R. J. Proc. Natl. Acad. Sci. USA. 1999;96:11693–11695. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gil J., Esteban M. Apoptosis. 2000;5:107–114. doi: 10.1023/a:1009664109241. [DOI] [PubMed] [Google Scholar]
- 12.Katze M. G., He Y., Gale M., Jr Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 13.Barber G. N. Cell Death Differ. 2005;12:563–570. doi: 10.1038/sj.cdd.4401643. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A., Haque J., Lacoste J., Hiscott J., Williams B. R. Proc. Natl. Acad. Sci. USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A., Yang Y. L, Flati V., Der S., Kadereit S., Deb A., Haque J., Reis L., Weissmann C., Williams B. R. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong A. H., Tam N. W., Yang Y. L., Cuddihy A. R., Li S., Kirchhoff S., Hauser H., Decker T., Koromilas A. E. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu W. M., Ostertag D., Li Z. W., Chang L., Chen Y., Hu Y., Williams B., Perrault J., Karin M. Immunity. 1999;11:721–731. doi: 10.1016/s1074-7613(00)80146-6. [DOI] [PubMed] [Google Scholar]
- 18.Cuddihy A. R, Li S., Tam N. W., Wong A. H., Taya Y., Abraham N., Bell J. C., Koromilas A. E. Mol. Cell. Biol. 1999;19:2475–2484. doi: 10.1128/mcb.19.4.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnet M. C., Weil R., Dam E., Hovanessian A. G., Meurs E. F. Mol. Cell. Biol. 2000;20:4532–4542. doi: 10.1128/mcb.20.13.4532-4542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deb A., Zamanian-Daryoush M., Xu Z., Kadereit S., Williams B. R. EMBO J. 2001;20:2487–2496. doi: 10.1093/emboj/20.10.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pflugheber J., Fredericksen B., Sumpter R., Jr, Wang C., Ware F., Sodora D. L., Gale M., Jr Proc. Natl. Acad. Sci. USA. 2002;99:4650–4655. doi: 10.1073/pnas.062055699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donze O., Deng J., Curran J., Sladek R., Picard D., Sonenberg N. EMBO J. 2004;23:564–571. doi: 10.1038/sj.emboj.7600078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gil J., Garcia M. A., Gomez-Puertas P., Guerra S., Rullas J., Nakano H., Alcami J., Esteban M. Mol. Cell. Biol. 2004;24:4502–4512. doi: 10.1128/MCB.24.10.4502-4512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel R. C., Sen G. C. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T., Yang M., May W. S. J. Biol. Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 26.Patel C. V., Handy I., Goldsmith T., Patel R. C. J. Biol. Chem. 2000;275:37993–37998. doi: 10.1074/jbc.M004762200. [DOI] [PubMed] [Google Scholar]
- 27.Ruvolo P. P., Gao F., Blalock W. L., Deng X., May W. S. J. Biol. Chem. 2001;276:11754–11758. doi: 10.1074/jbc.M011400200. [DOI] [PubMed] [Google Scholar]
- 28.Dar A. C., Dever T. E., Sicheri F. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 29.Dey M., Cao C., Dar A. C., Tamura T., Ozato K., Sicheri F., Dever T. E. Cell. 2005;122:901–913. doi: 10.1016/j.cell.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 30.Nanduri S., Carpick B. W., Yang Y., Williams B. R., Qin J. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanduri S., Rahman F., Williams B. R., Qin J. EMBO J. 2000;19:5567–5574. doi: 10.1093/emboj/19.20.5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters G. A., Hartmann R., Qin J., Sen G. C. Mol. Cell. Biol. 2001;21:1908–1920. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel R. C., Sen G. C. J. Biol. Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- 34.Tian B., Mathews M. B. J. Biol. Chem. 2001;276:9936–9944. doi: 10.1074/jbc.M007328200. [DOI] [PubMed] [Google Scholar]
- 35.Craig A. W., Cosentino G. P., Donze O., Sonenberg N. J. Biol. Chem. 1996;271:24526–24533. doi: 10.1074/jbc.271.40.24526. [DOI] [PubMed] [Google Scholar]
- 36.Hanks S. K., Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 37.Cai R., Williams B. R. J. Biol. Chem. 1998;273:11274–11280. doi: 10.1074/jbc.273.18.11274. [DOI] [PubMed] [Google Scholar]
- 38.Qin J., Vinogradova O., Cronenborn A. M. Methods Enzymol. 2001;339:377–389. doi: 10.1016/s0076-6879(01)39323-0. [DOI] [PubMed] [Google Scholar]
- 39.Galabru J., Hovanessian A. J. Biol. Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 40.Romano P. R., Green S. R., Barber G. N., Mathews M. B., Hinnebusch A. G. Mol. Cell. Biol. 1995;15:365–378. doi: 10.1128/mcb.15.1.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpick B. W., Graziano V., Schneider D., Maitra R. K., Lee X., Williams B. R. J. Biol. Chem. 1997;272:9510–9516. doi: 10.1074/jbc.272.14.9510. [DOI] [PubMed] [Google Scholar]
- 42.Wu S., Kaufman R. J. J. Biol. Chem. 1997;272:1291–1296. doi: 10.1074/jbc.272.2.1291. [DOI] [PubMed] [Google Scholar]
- 43.Romano P. R., Garcia-Barrio M. T., Zhang X., Wang Q., Taylor D. R., Zhang F., Herring C., Mathews M. B., Qin J., Hinnebusch A. G. Mol. Cell. Biol. 1998;18:2282–2297. doi: 10.1128/mcb.18.4.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vattem K. M., Staschke K. A., Zhu S., Wek R. C. Eur. J. Biochem. 2001;268:1143–1153. doi: 10.1046/j.1432-1327.2001.01979.x. [DOI] [PubMed] [Google Scholar]
- 45.Taylor S. S., Haste N. M., Ghosh G. Cell. 2005;122:823–825. doi: 10.1016/j.cell.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Ausubel F. M. New York: Greene and Wiley Interscience; 1988. Current Protocols in Molecular Biology. [Google Scholar]
- 47.Peters G. A., Khoo D., Mohr I., Sen G. C. J. Virol. 2002;76:11054–11064. doi: 10.1128/JVI.76.21.11054-11064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goh K. C., deVeer M. J., Williams B. R. EMBO J. 2000;19:4292–4297. doi: 10.1093/emboj/19.16.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nanduri S., Carpick B., Yang Y., Williams B. R., Qin J. J. Biomol. NMR. 1998;12:349–351. doi: 10.1023/a:1008259729432. [DOI] [PubMed] [Google Scholar]
- 50.Velyvis A, Vaynberg J., Yang Y., Vinogradova O., Zhang Y., Wu C., Qin J. Nat. Struct. Biol. 2003;10:558–564. doi: 10.1038/nsb938. [DOI] [PubMed] [Google Scholar]