Figure 1.
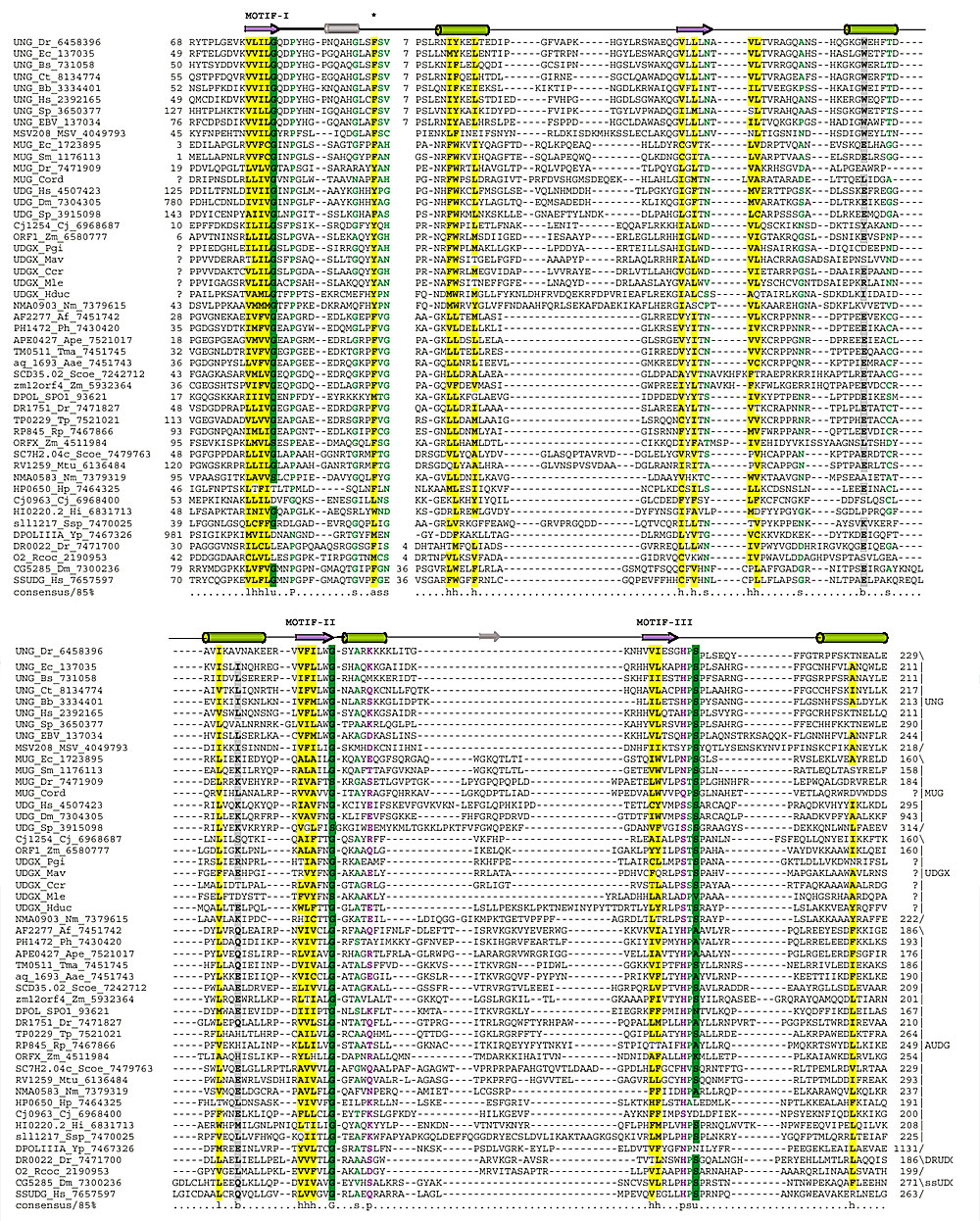
Multiple alignment of the UDG superfamily. The secondary-structure elements of the core UDG fold are shown in color above the multiple alignment. Some nonconserved elements in the MUG structure from E. coli are indicated in gray. The coloring of the alignment positions is according to the 85% consensus that includes the following categories of amino acid residues: h, hydrophobic, l, aliphatic, a, aromatic, shaded yellow (YFWLIVMA); s, small, individual letters colored green (SAGTVPNHD); p, polar, colored purple (STQNEDRKH); u, tiny, shaded green (GAS); and b, big, shaded gray (KREQWFYLMI). Af, Archaeoglobus fulgidus; Bb, Borrelia burgdorferi; Bs, Bacillus subtilis; Cj, Campylobacter jejuni; Ct, Chlamydia trachomatis; Dm, Drosophila melanogaster; Dr, Deinococcus radiodurans; Ec, Escherichia coli; Hi, Haemophilus influenzae; Hp, Helicobacter pylori; Hs, Homo sapiens; Mtu, Mycobacterium tuberculosis; Ph, Pyrococcus horikoshii; Rp, Rickettsia prowazekii; Sc, Saccharomyces cerevisiae; Scoel, Streptomyces coelicolor; Sp, Schizosaccharomyces pombe; Ssp, Synechocystis sp.; Tp, Treponema pallidum; Uu, Ureaplasma urealyticum; Yp, Yersinia pestis. The numbers at each end of each sequence are amino-acid positions and indicate the extent of the domain in each protein. The numbers within the alignment indicate inserts that have not been shown. The conserved motifs discussed in the text are designated I, II and III; the conserved aromatic (aliphatic) residue involved in the stacking interaction with uracil is indicated by an asterisk.
