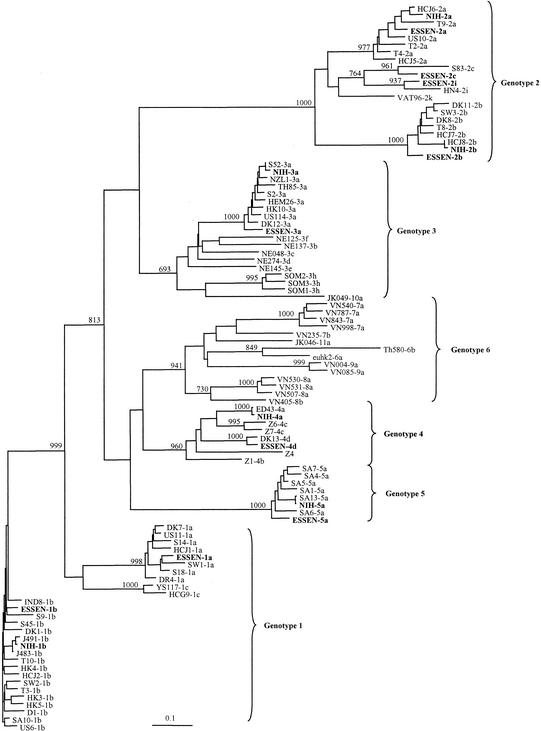
FIG. 2.
Phylogenetic analysis of E1 sequences generated from two HCV genotyping panels: the Essen panel (1a, 1b, 2a, 2b, 2c, 2i, 3a, 4, and 5a) and the NIH panel (1a, 2a, 2b, 3a, 4a, and 5a). The 83 reference sequences were collected from the literature as follows: D, DK, DR, HK, IND, S, SA, SW, T, US6, US10, US11, Z (6, 7); HC-G9, YS (20); HC-J1 (19); HC-J5, HC-J7, HC-J8 (21); HC-J6 (22); HEM, NZL, TH, US114 (22); NE (32); VN (34); VAT69 (31); JK046 and JK049 (36); Th580 (37); ED43 (8); HN4 (GenBank X76411) (29); and SOM (1). The phylogenetic tree was constructed by using the maximum-likelihood algorithm and the neighbor-joining method in the PHYLIP package (12). The numbers on the branching represent bootstrap 1000 values.