Table 1.
Differentially expressed transcripts found in Nb2 cells during cell-cycle progression using five different screening techniques
| Identity | Accession number | Expression variations | |
| Differential display | |||
| Unknown DD3 | No EST | Three transcripts (2, 2.8, 3.7 kb) | |
| expressed in G1 and G1/S, | |||
| (>4-fold induction) | |||
| Representational difference analysis: G1/S-GA | |||
| ATP synthase β subunit | M19044 | Induced in G1/S, G2 (2-fold induction) | |
| Aldehyde dehydrogenase | U79118 | Induced in late G1 (2-fold induction) | |
| Enolase α1 | NM012554 | Induced in late G1 (2-fold induction) | |
| Dynein heavy chain | D13896 | Induced in G1, G1/S (3-fold induction) | |
| TCP-1 ε | D43950(h) | Induced in G1/S, G2 (3-fold induction) | |
| αCOP (COPA) | U24105 | Induced in late G1 (2-fold induction) | |
| Cdc21 homolog | D26089 (m) | Peaks in G1/S (>8-fold induction) | |
| Ribosomal protein L26 | A1716351 | Induced in late G1 (2-fold induction) | |
| Itm1 | A1956728 (h) | Induced in G1/S (2-fold induction) | |
| Unknown T22 | A1053031 (h) | 6 kb, 2-fold induction in G1/S | |
| Unknown T34 | A1235326 | 2 kb, 2-fold induction in G1/S | |
| Subtractive suppressive hybridization: G1-GA | |||
| Galectin-8 | U09824 | Induced in G1 (2-fold induction) | |
| Hsp86 | X16857 | Induced in late G1 (>3-fold induction) | |
| TCP-1 η | AA900460 | Induced in late G1 (2-fold induction) | |
| Ribosomal protein L13A | X68282 | Induced in late G1 (2-fold induction) | |
| Ribosomal protein L12 | AA900142 | Induced in late G1 (2-fold induction) | |
| Ribosomal protein L3 | A1687295 | Induced in G1 (2-fold induction) | |
| Y00441 | Induced in G1 (2-fold induction) | ||
| β2-microglobulin | D12771 | Induced from late G1 to G2 (>5 fold induction) | |
| Adenine nucleotide translocator ANT-2 | NM011342 (m) | Induced in G1 (2-fold induction) | |
| Sec-22 | NM012562 | Induced in G1 (2-fold induction) | |
| L-fucosidase | U91538 | Induced in G1/S (>3-fold induction) | |
| CRM-1 homolog/exportin 1 | X81839 | Induced in G1/S (>3-fold induction) | |
| Ubiquitin/ribosomal protein S27a | A47416 | Induced in G1/S (>3-fold induction) | |
| Ubiquitin/ribosomal protein S30 (FAU) | AF195142 (m) | 3-fold induction in G1 (1.5 kb) | |
| Unknown 4-2 (mouse selenoprotein R mRNA) | H35219 | 2-fold induction in G1/S (2 and 4 kb) | |
| Unknown 4-4 (human KIAA0081) | No EST | 2-fold induction in G1 (1 kb) | |
| Unknown 4-11 | AU035826 | 4-fold induction in G1 (1 and 1.5 kb) | |
| Unknown 4-15 | AF046001 | 4-fold induction in G1 (2.5 kb) | |
| Unknown 4-16 (human ZNF207 or mouse Zep) | No EST | 3-fold induction in G1 (1.5 kb) | |
| Unknown 4-20 | AW435432 | 2-fold induction in G1 | |
| Unknown 4-27 (new ribosomal protein L15 type) | A1121996 (m) | 2-fold induction in G1 | |
| Unknown 4-49 | AW246248 (h) | 2-fold induction in G1 | |
| Unknown 4-58 (SH3, Rab GAP, TBC domain) | No EST | 2-fold induction in G1 | |
| Unknown 4-59 | |||
| Subtractive suppressive hybridization: GA-G1 | |||
| Spermidine/spermine N-acetyl transferase (SSAT) | AA955996 | Repressed transiently in G1 | |
| Leukocyte common antigen (alternative splicing) CD45 | Y00065 | Switch between two transcripts (one | |
| repressed, the other induced in G1) | |||
| ZFX | X75171 | Repressed transiently in G1 | |
| Ribosomal protein S8 | AA874997 | Repressed transiently in G1 | |
| Ribosomal protein S13 | L01123 | Repressed transiently in G1 | |
| Unknown 6-2 | No EST | Repressed transiently in G1 | |
| Unknown 6-3 (hypothetical protein expressed in thymocytes) | AJ237585 (m) | Repressed transiently in G1 | |
| Unknown 6-4 | No EST | Repressed in G1 | |
| Unknown 6-9 | No EST | Repressed transiently in G1 | |
| Unknown 6-10 | No EST | Repressed transiently in G1 | |
| Unknown 6-12 (human protein KIAA0710) | AB014610 (h) | Repressed transiently in G1 | |
| Unknown 6-45 (homolog to mouse PARP-2) | NM009632 (m) | Repressed transiently in G1 | |
| Differential screening of a rat organized library: G2-GA | |||
| Prothymosin α | M86564 | Induced in G2 | |
| Cyclophilin | M19533 | Induced in G2 | |
| ATP synthase β subunit | M19044 | Induced in G1/S and G2 | |
| Tubulin α2 | AA686718 | Induced in G1/S and G2 | |
| GaPDH | X02231 | Induced in G2 | |
| Phosphoglycerate kinase | M31788 | Induced in G1/S and G2 | |
| MRG1 related protein | U65093 | Induced in G1/S and G2 | |
| Unknown BO1 | No EST | Induced in G2 | |
| Analysis of weakly expressed candidate genes: UN, A, GI, G1/S, G2 | |||
| Name | Accession number | Fold induction | Kinetics |
| Ganglioside synthase (GD3) | D84068 | > 4 | 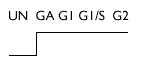 |
| P13 kinase | D64045 | > 3 | |
| Phospholipase Cγ1 | M34667 | > 3 | |
| Bax | S76511 | > 2 |  |
| P53 | X13058 | > 2 | |
| FAKp125 | AF020777 | > 3 | |
| 14-3-3 ε | M84416 | > 2 | |
| 14-3-3 η | D17445 | > 2 | 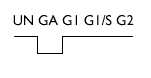 |
| Vitamin D3 receptor | J09838 | 3 | |
| Glycine transporter | M88595 | 3 |  |
| Thromboxane A2 receptor | D32080 | 2 | 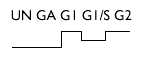 |
| Phosphatidylinositol transfer protein | D17445 | 2 | 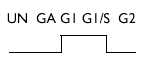 |
| RexB/NSP | U17604 | > 2 |  |
| Glucocorticoid receptor | M14053 | 2 | 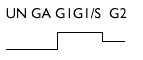 |
| Ga3PDH | J04147 | 2 | 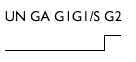 |
| Zif268 = EGR1 | U75398 | 2 |  |
Nb2 cells: UN, unsynchronized; GA, growth arrested; G1, G1 phase; G1/S, G1/S transition; G2, G2 phase. PC12, rat pheochromocytoma PC12 cells. 18S and 28 S, ribosomal RNA. Whenever possible, rat accession numbers (GenBank) are written in the second column. When this sequence is not in known for the rat, (h) or (m) indicates that the accession number corresponds, respectively, to a human and a mouse cDNA homologous to those of the rat. `No EST' means that our rat sequence does not correspond to any previously described EST in mammals. For some of the unknown cDNAs, an estimation of the size of the corresponding transcript(s) as well as the fold induction is given in the right column `Expression variations', and for weakly expressed candidate genes, the expression kinetics are shown.
