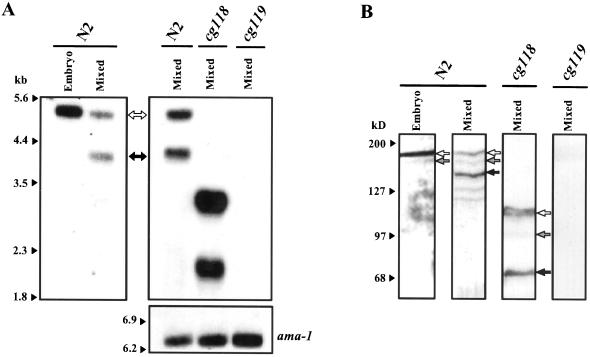
Figure 3.
Northern and Western blot analyses of nid-1 expression in wild-type and mutant animals. (A) Detection of nid-1 transcripts in RNA extracted from wild-type (N2) and nid-1 mutant (cg118, cg119) animals. RNAs were extracted from embryos (Embryo) or populations of animals of mixed development stages (Mixed). The positions of nid-1A (open arrow) and nid-1C (filled arrow) transcripts are indicated between A and B. This blot was probed with radiolabeled nid-1 exon 15 to reveal all isoforms. Equivalent blots were also probed with exons 11 and 13. Hybridization with the RNA polymerase II gene (ama-1) was used as a loading control. The positions of size standards are indicated on the left. (B) Detection of NID-1 in extracts from wild-type (N2) and nid-1 mutant (cg118, cg119) animals. Extracts were made from embryos (Embryo) or populations of animals of mixed development stages (Mixed). The positions of putative NID-1A (open arrows), NID-1B (shaded arrows), and NID-C (filled arrows) are indicated to the right of each lane. The positions of molecular weight standards are indicated on the left.