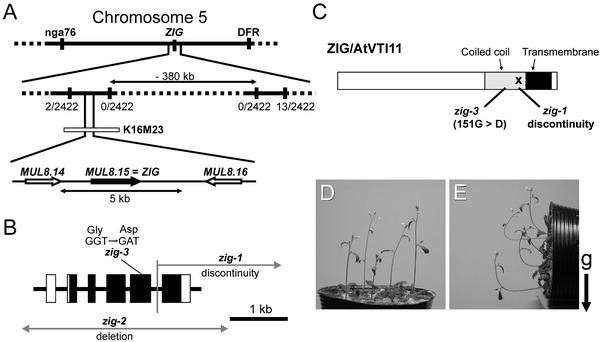
Figure 8.
Molecular Cloning of the ZIG Gene.
(A) Initial mapping placed the zig mutation between the nga76 and DFR markers. Using newly generated PCR markers, the position of the ZIG locus was narrowed to an ∼380-kb region. The numbers of recombinants among 2422 chromosomes tested are indicated (recombinant chromosomes/analyzed chromosomes). DNA gel blot analysis using the K16M23 TAC clone as a probe showed that deletions had occurred in both zig-1 and zig-2 in the region that contains MUL8.15.
(B) The structure of the ZIG gene and the mutation sites of the zig alleles examined. Boxes represent exons: closed boxes, translated regions; open boxes, untranslated regions. zig-2 had lost the ZIG gene completely as well as some neighboring genes (data not shown).
(C) Molecular lesion in each allele shown on the protein. The X shows the chromosomal discontinuity in zig-1. Black and gray boxes indicate a predicted transmembrane domain and a coiled-coil domain, respectively, of ZIG/AtVTI11.
(D) and (E) Complementation of zig by the wild-type ZIG genomic region. The 5-kb genomic DNA fragment that includes MUL8.15 (A) was transformed into zig-1 plants.
(D) The resulting 4-week-old transgenic plants.
(E) Plants placed horizontally for 2 hr at 23°C in the dark. The arrow indicates the direction of gravity (g).