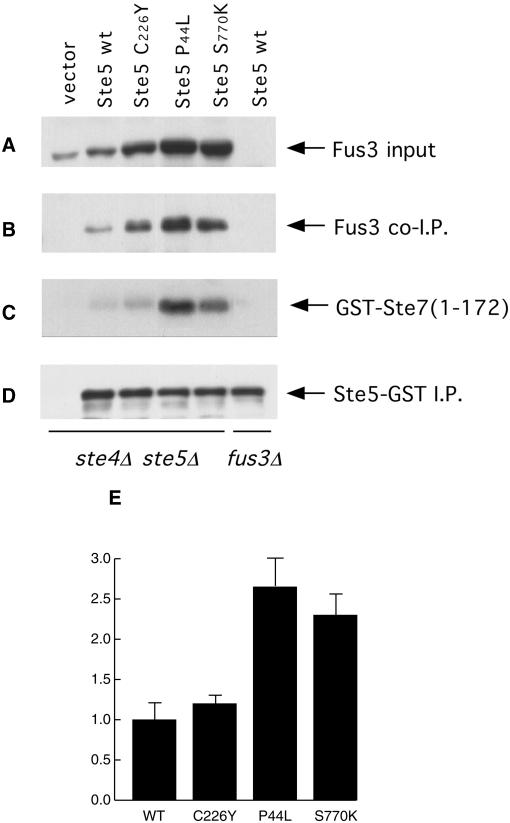
Figure 6.
Association of activated Ste5 mutants with Fus3. A ste4Δ ste5Δ strain was transformed with a vector or with the same vector expressing under the control of the GAL1 promoter either wild-type STE5-GST or the three indicated STE5-GST mutants. The cells were grown, induced with galactose, and extracts were prepared, as described in the legend to Figure 5. The c-Myc-tagged Ste5-GST proteins were immunoprecipitated (D) and a portion of the bead-bound immune complexes was analyzed as also described in the legend to Figure 5. The amount of endogenous Fus3 in a sample of each extract (“input”) (A) and the amount of endogenous Fus3 that coimmunoprecipitated with Ste5 (B) were assessed by immunoblotting with anti-Fus3 antibodies. (C) A separate portion of the bead-bound immune complexes was washed three times with lysis buffer and twice with kinase assay buffer, and then Fus3 activity present was measured by the incorporation of label into a specific substrate, GST-Ste7(1–172), as described in MATERIALS AND METHODS. (E) Densitometric analysis was performed on the bands in B and C by using NIH imaging software. Values expressed are the ratio of the radiolabel incorporated into substrate in the Fus3 kinase assay (C) to the intensity of the Fus3 protein present in the same coimmunoprecipitates (B), are the average of three independent trials (error bars represent SD of the mean), and are normalized to the value obtained for the immunoprecipitates containing wild-type Ste5.