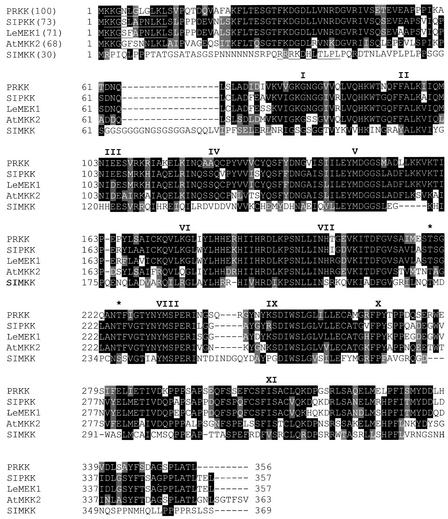
Figure 1.
Primary Structure of PRKK, a Pathogen-Responsive Alfalfa MAPKK.
The amino acid sequence of PRKK was aligned with its closest homologs belonging to the PMKK1 subfamily of plant MAPKKs (Ligterink and Hirt, 2001). PRKK showed 73% identity to tobacco SIPKK (Liu et al., 2000), 71% identity to tomato LeMEK1 (Hackett et al., 1998), and 68% identity to Arabidopsis AtMKK2 (Ichimura et al., 1998a). For comparison, SIMKK also was included (Kiegerl et al., 2000). Identical and conserved amino acids are shaded in black and gray, respectively; dashes represent gaps. The 11 catalytic subdomains are represented by roman numerals above the respective regions, and amino acids defining the putative MAPK docking site are underlined. The putative phosphorylation sites are indicated by asterisks; note that the consensus sequence for plant MAPKKs in this region is S/T-(X)5-S/T and not S/T-(X)3-S/T, as in yeast and animal MAPKKs (Alessi et al., 1994).