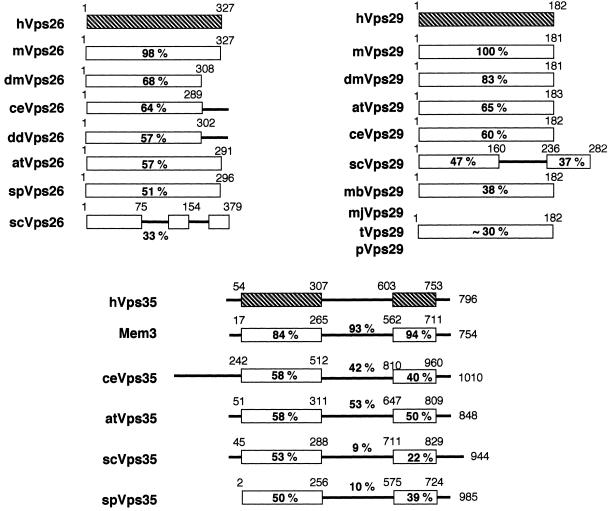
Figure 2.
Human Vps26, Vps29, and Vps35 are evolutionarily well conserved. Shown are schematic illustrations of the predicted domain architecture of human (h) Vps26, Vps29, and Vps35, and related proteins from C. elegans (ce), S. cerevisiae (se), Schizosaccharomyces pombe (sp), mouse (m), A. thaliana (at), D. melanogaster (dm), Dictyostelium discoideum (dd), M. jannaschii (mj), M. thermoautotrophicum (mb), T. maritima (t), and P. abyssi (p). The accession numbers of the homologous molecules are as follows: mVps26, P40336; dmVps26, T12683, ceVps26, O01258; ddVps26, AAB03668; atVps26, T05874; spVps26, Q10243; scVps26, NP004887; mVps29, AF193794; dmVps29, AAF51410; atVps29, T07720; ceVps29, T27697; scVps29, NP011876; mjVps29, Q58040; mbVps29, O27802; tVps29, D72296; pVps29, H75143; Mem3, NP032608, ceVps35, T34314; atVps35, AAB80794; scVps35, NP012381; spVps35, T11719. The percentage of amino acid identity between each human protein and related proteins from other species as determined by CLUSTAL W alignment program (Thompson et al., 1994) is shown below the various domains of the proteins.