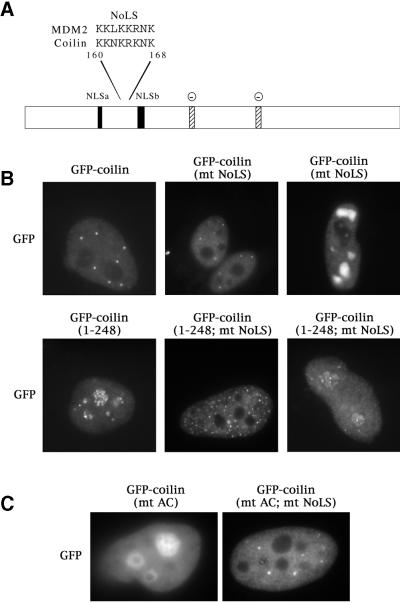
Figure 4.
Identification and mutation of a nucleolar localization signal (NoLS) in coilin. (A) Coilin contains a sequence, located between the two putative NLSs, that is nearly identical to the NoLS of MDM2 (Lohrum et al., 2000). Stippled boxes indicate potentially-acidic serine patches (AC) spanning amino acids 242–259 and 312–325. Facing page: (B) The putative coilin NoLS was mutated from KKNKRKNK to IINNIINI (mt NoLS) in both the GFP-coilin and GFP-coilin(1–248) backgrounds. Note that, while the truncation alone localized to nucleoli and CBs, mutation of the NoLS depleted coilin from nucleoli. (C) Similarly, block substitution of the two acidic patches (S245–252A and S314–320A, termed GFP-coilin(mt AC)) also resulted in nucleolar accumulation. Additional localization in CBs was occasionally observed (our unpublished results). However, mutant proteins bearing substitutions in both the NoLS and the acidic patches (GFP-coilin(mt AC; mt NoLS)) did not accumulate in nucleoli, resulting in a distribution pattern indistinguishable from that of wild-type coilin.