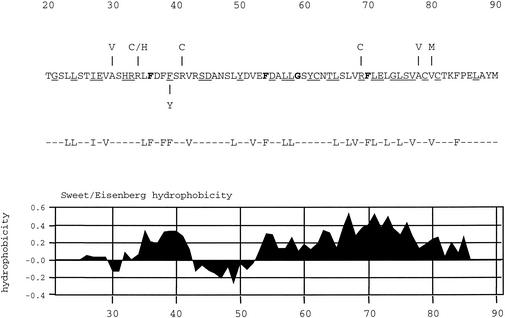
FIG. 1.
Amino acid sequence of the N terminus of VP5, which is important for interaction with the scaffold protein. The amino acid sequence of VP5 (strain KOS) from residues 20 to 90 is shown. Displayed above the sequence are the mutations identified in the revertant viruses that overcame the blocked maturation cleavage site in the scaffold proteins (36). Also illustrated below the sequence is the most frequent mutation (F39→Y) discovered in wild-type VP5, which reduced the VP5-pre-22a interaction in the yeast two-hybrid assay (37). The 24 hydrophobic residues targeted for mutation are shown below this figure. All the residues were changed to alanine. Displayed below the targeted amino acids is a plot of the hydrophobicities of the amino acids in this sequence, as determined with the Sweet-Eisenberg computer program (32). Residues in bold are conserved in all herpesviruses examined (herpes simplex viruses type 1 and type 2, pseudorabies virus, equine herpesvirus 1, bovine herpesvirus 1, varicella-zoster virus, turkey herpesvirus 2, human herpesvirus 8, equine herpesvirus 2, Epstein-Barr virus, murine herpesvirus 68, alcelaphine herpesvirus 1, human herpes virus 6, human herpesvirus 7, human cytomegalovirus, simian cytomegalovirus, and murine cytomegalovirus. The residues that are underlined are conserved in the alphaherpesviruses (herpes simplex viruses type 1 and type 2, pseudorabies virus, equine herpesvirus 1, bovine herpesvirus 1, turkey herpesvirus 2, and varicella-zoster virus).