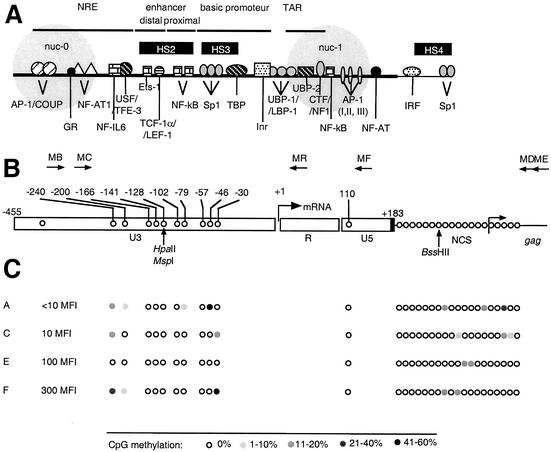
FIG. 1.
CpG methylation pattern of the LTR region in LTR-GFP clonal cell lines expressing different levels of GFP. (A) Transcription elements in the HIV-1 LTR. NRE, negative regulatory element; nuc-0 and nuc-1, nucleosomes. Black boxes, HS, DNase I-hypersensitive sites. (B) Positions and orientations of PCR primers (MB, MC, MD, ME, MF, and MR) used to amplify bisulfite-treated HIV-1 DNA. The arrow at the U3-R junction denotes the start site of transcription (nucleotide +1). Positions of CpG dinucleotides within the HIV-1 LTR, the adjacent leader region, and the coding region of Gag are shown by open circles. Positions are noted with respect to nucleotide +1. NCS, noncoding sequence. Positions of restriction endonucleases sensitive to CpG methylation are shown by vertical arrows. (C) Positions of CpG dinucleotides within the HIV-1 LTR are shown by open circles and noted with respect to nucleotide +1. Open circles, nonmethylated CpG residues; closed circles, methylated CpG residues, grey scale indicates the percentage of CpG methylation. The MFIs of clonal cell lines A, C, E, and F are shown. Low basal activity is indicated by an MFI of <0.15, and a high basal activity is indicated by an MFI of >100.