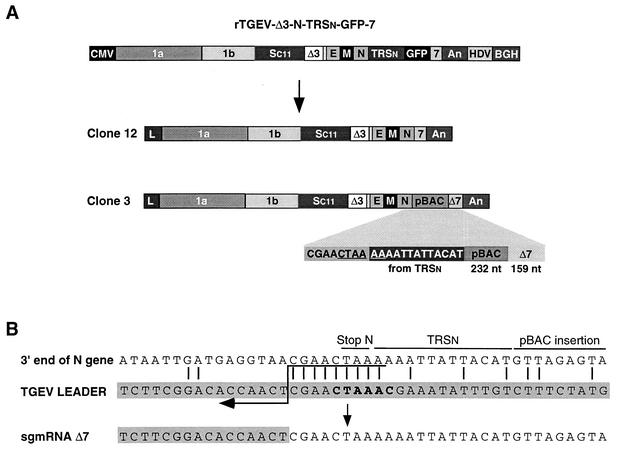
FIG. 8.
Genetic structure of recombinant viruses derived from rTGEV-Δ3-N-TRSn-GFP-7. (A) Schematic structure of the cDNA encoding rTGEV-Δ3-N-TRSn-GFP-7 RNA and the resulting viral clones 12 and 3, showing the new organization at the 3′ end of the genome. Below the shaded area of clone 3, the last nucleotides of the N gene, the 5′-most 13 nucleotides of TRSn, a 232-nucleotide insertion derived from the pBAC plasmid, and the 3′-end 159 nucleotides of gene 7 are represented. The noncanonical core sequence generated at the 3′ end of the N gene is underlined. The numbers and letters inside the rectangles indicate the viral genes. CMV, cytomegalovirus immediate-early promoter; An, poly(A); HDV, hepatitis delta virus ribozyme; BGH, bovine growth hormone termination and polyadenylation signals. pBAC, insertion derived from the pBAC plasmid. Δ7, gene 7 with a 78-nucleotide deletion at the 5′ end. (B) Nucleotide identity between the leader RNA of TGEV and the genomic sequences of rTGEV-Δ3-N-TRSn-GFP-7 clone 3 flanking the noncanonical junction site at the 3′ end of the N gene that led to sgmRNA Δ7. Letters above genomic sequences indicate the origin of nucleotides. The leader region (shaded nucleotides) includes the canonical core sequence (in bold type) and the 20 immediately upstream and downstream nucleotides. Leader-body common nucleotides are indicated by vertical bars. Postulated polymerase strand switching during minus-strand synthesis is indicated by the arrow. The jump during discontinuous transcription could have occurred anywhere within the consecutive nucleotides underlined by the tail of the arrow.