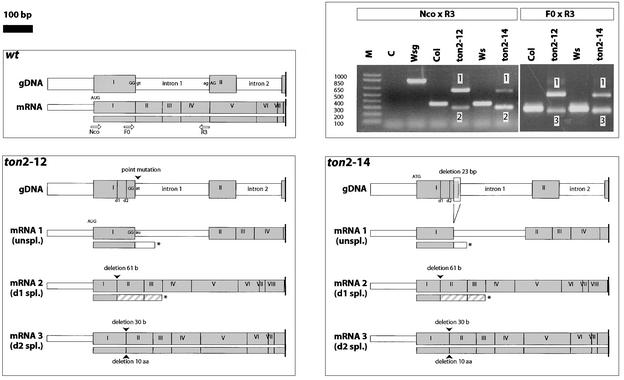
Figure 4.
Molecular Defects in ton2 Weak Alleles ton2-12 and ton2-14.
The structures of genomic DNA (gDNA) and transcripts (mRNA) are represented for wild-type, ton2-12, and ton2-14 alleles. For clarity, only the 5′ part of the ton2 locus is indicated. Gray boxes represent exons (numbered I to VIII), and white boxes represent introns. The predicted structure of the encoded polypeptide is indicated under each transcript (gray indicates an in-frame translation product of exons; hatched gray indicates out-of-frame translation of exons; white indicates translation of introns; and the asterisk indicates the stop codon). The positions of RT-PCR primers are indicated by white arrows. The positions of alternative donor sites d1 and d2 are indicated. Sequencing of genomic DNA revealed that both weak alleles have a mutation affecting the donor site of intron 1. ton2-12 has a single-base change that converts this site from TGGgt to TGGat, and ton2-14 has a 23-bp deletion that removes the same splice site. RT-PCR analyses of wild-type and weak alleles are shown at top right. All transcripts were characterized by RT-PCR and sequencing of RT-PCR products. As a result of the mutations affecting the donor site of intron 1, splicing of this intron is perturbed and three transcripts are detected in each mutant (bands 1, 2, and 3). In both mutants, two major transcripts were revealed by RT-PCR using primers TON2Nco and TON2R3. Transcript 1 is unspliced and contains intron 1. Transcript 2 is misspliced (d1 donor site) and has the last 61 nucleotides of exon I deleted. These two abnormal transcripts are expected to give nonfunctional truncated polypeptides comprising (part of) exon I with small extensions. However, in both weak mutants, an alternative misspliced transcript (mRNA 3, d2 donor site) of lower abundance can be detected using other RT-PCR primers (Ton2F0 and Ton2R3) that are not able to amplify transcript 2. This second misspliced transcript contains a 30-nucleotide in-frame deletion at the end of exon I, resulting in a 10–amino acid truncation (Gly-38 to Met-47) in the predicted translation product. aa, amino acids; C, control (reverse transcriptase omitted); M, molecular mass markers; spl., spliced; unspl., unspliced; Wsg, genomic DNA; wt, wild type.