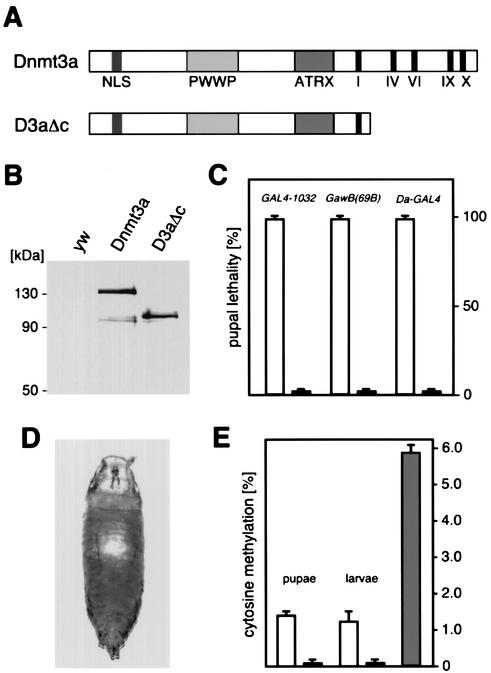
FIG. 1.
Expression of mouse DNA methyltransferase Dnmt3a in D. melanogaster. (A) Dnmt3a constructs used for GAL4-dependent overexpression. The conserved catalytic motifs of the C-terminal catalytic domain are shown as black bars. NLS indicates the predicted nuclear localization signal. The PWWP domain has been implicated in DNA binding (52), and the ATRX domain has been implicated in histone deacetylase interactions (19). (B) Equal expression from all transgenes was confirmed by Western blotting. No protein was detectable in control crosses (yw). (C) Expression of Dnmt3a transgenes with various GAL4 lines that result in high-level and widespread transactivation. Expression of the full-length transgene caused lethality in all of the crosses analyzed (open bars), and no phenotype could be observed upon overexpression of the control construct (black bars). (D) Appearance of the DNA hypermethylation-induced phenotype after Da-GAL4 transactivation. (E) Genomic cytosine methylation levels were determined by capillary electrophoretic analysis. Dnmt3a overexpression with Da-GAL4 resulted in 1.4% cytosine methylation in early pupae and 1.2% cytosine methylation in third-instar larvae (open bars). Only background levels of cytosine methylation were detectable in control pupae and larvae overexpressing the D3aΔc control protein (black bars). Normal calf thymus DNA was included as an additional control (grey bar).