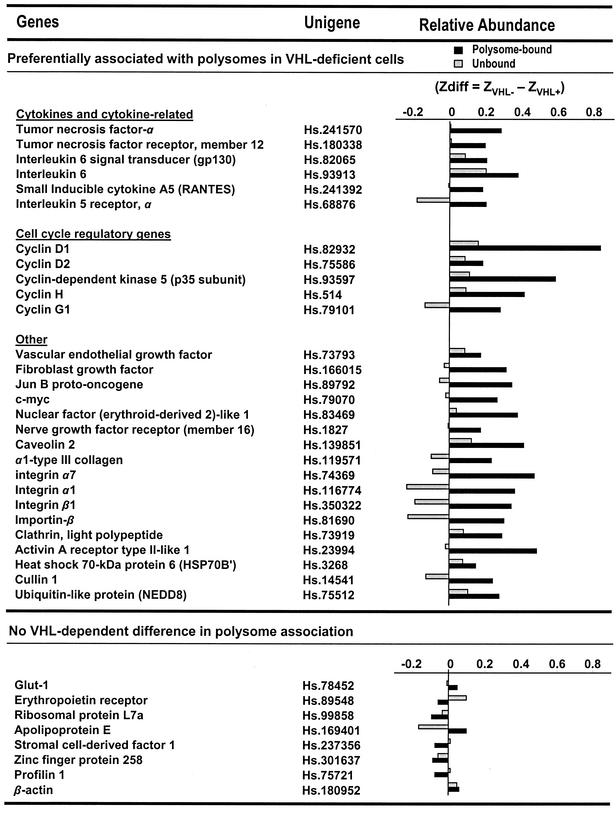
FIG. 4.
Collection of genes encoding mRNAs that are specifically bound to polysomes in pVHL-deficient cells. For each gene on the cDNA arrays, the relative presence of the encoded mRNA in polysome-bound and unbound fractions was compared between pVHL-expressing and pVHL-deficient 786-0 cell lines. Cell fractionations and cDNA array analysis were performed three independent times. Differences in Z averages (Zdiff) served to assess the relative abundance of a given transcript in polysomes (polysome bound; black bars) or in unbound fractions (gray bars) of pVHL-deficient and pVHL-expressing cells and were calculated as explained in Materials and Methods and in reference 5. The top group(preferentially associated with polysomes in pVHL-deficient cells) includes genes potentially subject to translational repression by pVHL, since they are encoded by mRNAs that were significantly more abundant in polysome-bound fractions prepared from pVHL-deficient cells than in those prepared from pVHL-expressing cells. *, Zdiff values for the polysome-bound mRNA are significant, using two tests (Z ≥ 2.4; P ≤ 0.05). For these genes, unbound mRNAs were not significantly different between pVHL-expressing and pVHL-deficient populations. The bottom group (no pVHL-dependent difference in polysome association) lists genes encoding mRNAs that exhibited no significant differences in their relative distribution (polysomal and unbound) in cells with different pVHL status.