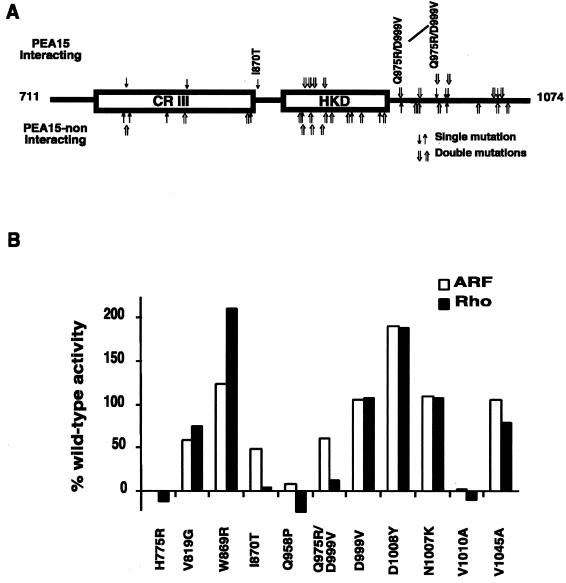
Figure 1.
Identification of two candidate RhoA-nonresponsive PLD1 mutants. (A) Distribution of single- and double-point mutations in the RhoA-noninteracting D4 fragments recovered from the split-hybrid screen and scoring of their interaction with PEA15. Arrows above the schematic diagram indicate alleles that continued to successfully interact with PEA-15 (H775R, V819G, I870T, N905Y/K981N, T906S/Q1046H, N913/D1008N, G921E/K1039E, Q975R/D999V, D999V, N1007K, V1010A, and V1045A); arrows below the schematic diagram indicate alleles that did not interact with PEA-15 (I770 M, H775L/E1054G, I777N, I805T, V817E/S979R, N867K/D980V, W869R, D903V/R1032P, D904N, D904G/A912T, A912G/L964R, K922 M/S939L, S925R/V1001N, M941T, A948V/V1045 M, Q958P, D976V, and D1008Y). Only the C-terminal D4 fragment of PLD1 (amino acids 711-1074) is shown. Depicted within it is conserved region III (amino acids 743–864) and the HKD domain (amino acids 888–965), both of which are critical for catalytic functioning independent of RhoA stimulation. The mutants I870T and Q975R/D999V are discussed further in the text. (B) Wild-type PLD1, PLD1-K898R (a catalytically inactive allele) (Sung et al., 1997), and candidate nonresponsive alleles recovered from the split-hybrid screen were overexpressed in COS-7 cells. The transfected cells were lysed and assayed for PLD activity in the presence of GTPγS-activated ARF1 and RhoA using the in vitro head-group release assay. Activities are shown in comparison to those exhibited by wild-type PLD1 and by PLD1-K898R, which define the 100% and 0% activities, respectively. Each mutant was assayed in duplicate a minimum of twice. Note that the PLD1-K898R behaves as a null allele, not as a dominant negative one, in both the in vitro and in vivo PLD assays (Zhang et al., 1999).