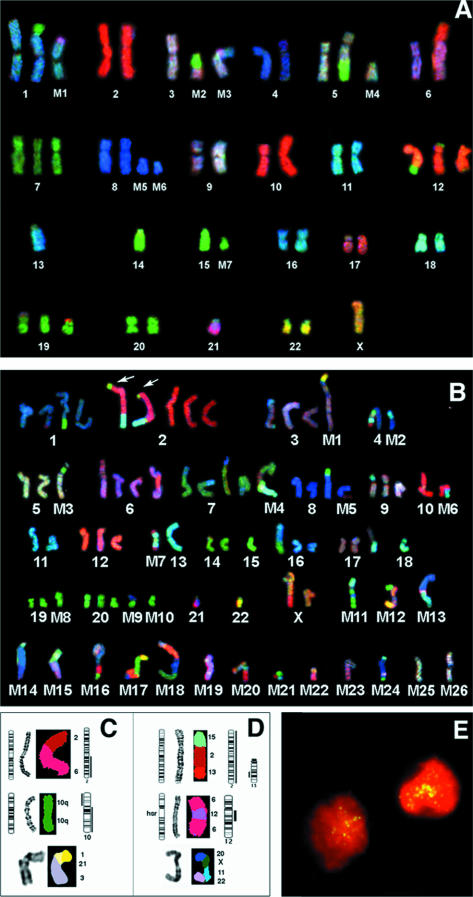
Figure 4.
SKY karyotypes for RD (A) and SJRH30 (B) are shown as RGB display, and individual rearrangements (panel C and D) are shown as classified SKY image display. A. SKY analysis of the embryonal RMS cell line RD. Identification is presented in Table 4 (Panel A). B. SKY analysis of the RMS-A cell line SJRH-30 Identification is presented in Table 4 (Panel B). C. Comparison between CGH and SKY analysis for RD. The schematic ideogram indicates the structural rearrangement based on a combination of SKY and G-banding. CGH profiles with loss on the left and gain on the right have been placed adjacent to each chromosome ideogram involved. For RD, the topmost rearrangement is an unbalanced 2;6 translocation, leading to 2q gain detected by CGH. The middle panel shows an isochromosome 10q with a gain of 10q and loss of 10p. The lower panel depicts a complex rearrangement detected by SKY observed in RD (M3 in Figure 3A and 4A). The chromosomes involved were 1, 21, and 3. D. Comparison between CGH and SKY for SJRH-30. The upper rearrangement is between regions of chromosomes 2, 13, and 15. This structural aberration has resulted in a partial loss and gain of the chromosomal segments involved as there is total gain of chromosome 2, and loss of part of the rearranged segment of distal 13q (also seen arrowed in panel B). The middle rearranged chromosome is an hsr-bearing chromosome 6, with amplified material mapping to the Gli and CDK4 region of chromosome 12 as identified by CGH. The lower panel displays a complex SKY chromosome marker (M12 on Figures 3B and 4B) with small segments of chromosomes involved (20, X, 11, 22). E. Interphase FISH analysis of a RMS-A tumor with DDX1 identifies gene amplification as dispersed ‘speckling’ of FISH signals within nuclei. This pattern of gene amplification is usually associated with the presence of dmin. The patient (A7 in Table 3) had 20 copies of MYCN and DDX1, which was confirmed by Southern blot analysis.