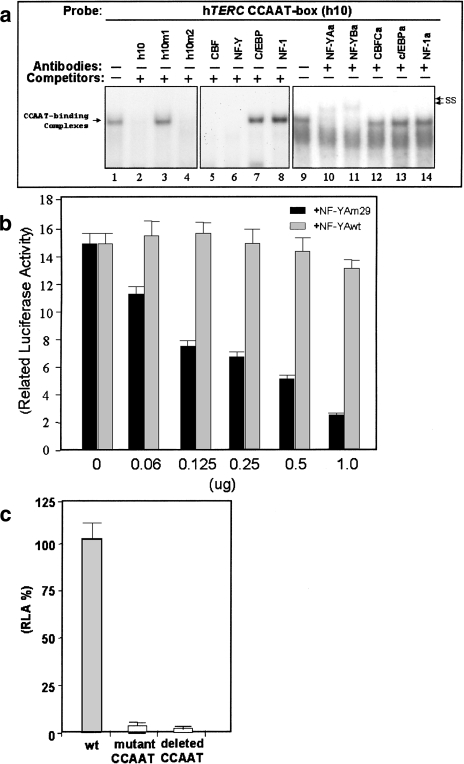
Figure 2.
(a) Identification of nuclear proteins binding to the CCAAT box. HeLa nuclear extract was mixed with radiolabeled oligonucleotide probe and analyzed by EMSA. Specific DNA-protein complexes are indicated by the arrow on the left. Complexes supershifted by preincubation with antibodies are shown on the right by arrows marked “ss.” The oligonucleotide h10 was used as a probe and the oligonucleotides used as competitors are indicated at the top of lanes 2 to 8. The antibodies used in supershifts are indicated above lanes 9 to 14. (b) Inhibition of hTERC promoter activity by a dominant-negative mutant of NF-YA termed NF-YAm29. 5637 cells were transfected with fixed amount of the hTERC-luciferase plasmid hProm176 and increasing concentrations of the NF-YAm29 dominant-negative vector (black columns) or wild-type NF-YA vector (light grey columns). For each transfection the mean and standard deviation for duplicate samples is shown. (c) Functional analysis of the CCAAT box. Promoter activities of a mutant CCAAT box construct (hProm176h10m1) and shorter promoter construct with deletion of the CCAAT box (hProm120) were assayed by transfection into the bladder carcinoma cell line 5637 and compared to the wild-type promoter activity (hProm176). Promoter activities of the constructs are shown as percentage luciferase activity of the wild-type promoter alone. The pSEAP2-Control vector was used as an internal control for transfection efficiency.