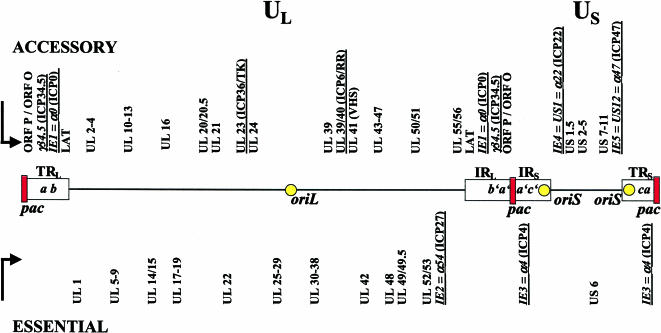
Figure 1.
HSV-1 genome structure. The virus genome is a linear, double-stranded DNA of ∼152 kb which encodes more than 80 genes. The genome is composed of unique long (UL) and unique short (US) segments, which are flanked by inverted repeats (R). IRL, internal repeat of the long segment; TRL, terminal repeat of the long segment; IRS, internal repeat of the short segment; TRS, terminal repeat of the short segment. The repeats of the L component are designated ab and b′a′; those of the S segment are a′c′ and ca. Pac signals are contained in the a sequences located at the junction between the long and short segments and at both termini. The HSV-1 genome contains two different origins of DNA replication, oriS and oriL. OriS is duplicated because it is located within the inverted repeats flanking US between the promoters of the IE3 and IE4/5 genes. OriL is located within UL and is flanked by transcriptional start sites of two E genes, which encode the single-stranded DNA binding protein, ICP8 (UL29), and the DNA polymerase (UL30). Approximately half of the genes are essential for virus replication in cell culture. The other half encode accessory functions, which contribute to the virus life cycle in specific tissues or cell types, e.g., postmitotic neurons. However, it can be assumed that the genes known to be dispensable for growth in cultured cells may be important for both optimal lytic replication and replication in vivo, contributing to pathogenesis, host range, latency, or spread in neurons. Genes underlined mark IE genes or genes which are relevant in certain recombinant HSV-1 mutants (see Part II).