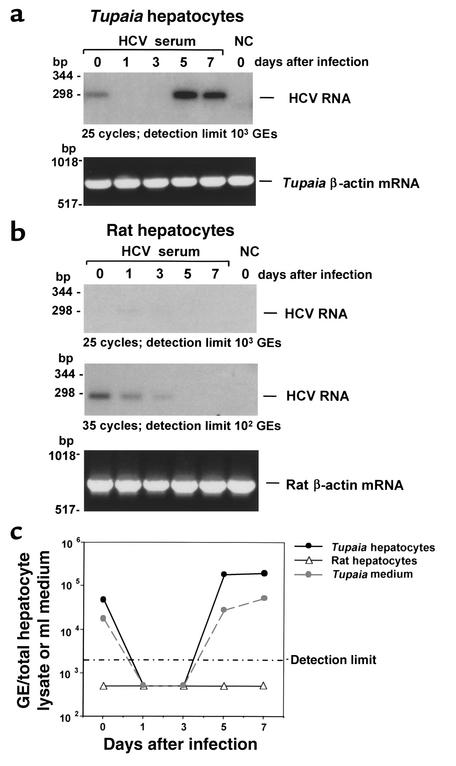
Figure 1.
(a) HCV infection of primary Tupaia hepatocytes. Primary Tupaia hepatocytes were incubated with 50 μl HCV RNA–positive serum 1 day after plating. At various timepoints after plating (days 0–7, as indicated), hepatocyte RNA was isolated, and positive-strand HCV RNA was analyzed by strand-specific RT-PCR using HCV-specific primers as described in Methods. The negative control (NC) consisted of primary Tupaia hepatocytes incubated with HCV-negative control serum harvested and analyzed on day 0 (day of incubation). To control for RNA integrity, cellular Tupaia β-actin mRNA was amplified by RT-PCR as described in Methods. The detection limit of the RT-PCR (25 cycles) was approximately 103 GEs/assay. (b) Lack of HCV infection in primary rat hepatocytes. Primary rat hepatocytes were incubated with the same HCV RNA–positive serum described in a, and HCV RNA was analyzed as described above. To control for RNA integrity, cellular rat β-actin mRNA was amplified by RT-PCR. The detection limit of the RT-PCR was approximately 103 GEs/assay when 25 cycles were used, and increased to approximately 102 GEs/assay when 35 cycles were used. (c) Quantitative analysis of HCV RNA synthesis in primary Tupaia and rat hepatocytes. Primary Tupaia and rat hepatocytes were incubated with HCV RNA–positive serum as described above. At various timepoints, positive-strand HCV RNA was quantified using the Amplicor HCV Monitor assay from Roche Diagnostic Corp. Cellular and medium HCV RNA levels are indicated as GEs/ml of hepatocyte lysate and medium. The detection limit of this assay was 2 × 103 GEs/ml.