Figure 5.
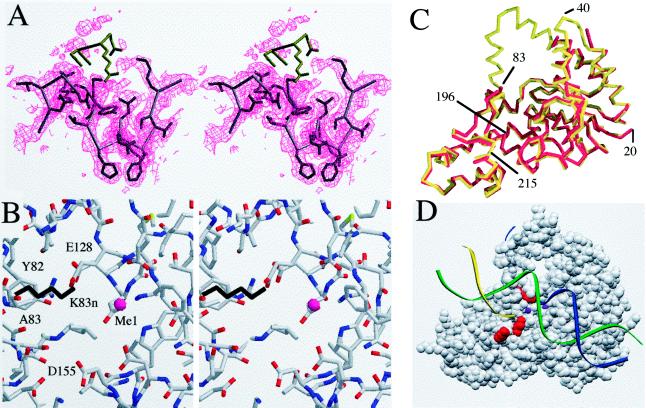
(A) Electron density map of the region around the mutated residue K83. The density for this residue is defined in a 2Fo − Fc map contoured at 1σ. The positions of the next few residues of the native molecule are colored in brown, and these residues are disordered. The next residue is G84; this position is expected to show flexibility. The helical arch does not show evidence of order in the electron density map. The first well defined residue in helix 5 is F104. The absence of density preceding this residue suggests that the helix is unwound and exists in an undefined conformation. (B) A stereo view of the metal-binding site MeI as determined in the native molecule. The K83 side chain from the native molecule is shown in black. The positions of the metal’s ligands are largely unchanged compared with the native (data not shown). The only amino group in close proximity to MeI is that of K83 (distance between NH2 and MeI = 4.2 Å). (C) A Cα trace of the native (red) and K83A mutant structures (yellow) superimposed. Disordered residues in the mutant include 35–41 and 84–103. The regions around the acidic residues composing the metal-binding site are very similar. (D) A space-filling model of the protein, showing the three residues mutated in this study in red (top to bottom: K83, K196, and K215) in relation to the proposed threading mechanism. DNA is shown as colored strands and the metal sites observed are shown as purple spheres.