Abstract
Long interspersed elements (LINE-1 or L1) are the most active transposable elements in the human genome. Due to their high copy number and ability to sponsor retrotransposition of nonautonomous RNA sequences, unchecked L1 activity can negatively impact the genome by a number of means. Substantial evidence in lower eukaryotes demonstrates that the RNA interference (RNAi) machinery plays a major role in containing transposon activity. Despite extensive analysis in other eukaryotes, no experimental evidence has been presented that L1-derived siRNAs exist, or that the RNAi plays a significant role in restricting L1 activity in the human genome. This review will present evidence showing a direct role for RNAi in suppressing the movement of transposable elements in other eukaryotes, as well as speculate on the role RNAi might play in protecting the human genome from LINE-1 activity.
IMPORTANCE OF LIMITING L1 RETROTRANSPOSITION IN THE HUMAN GENOME
The majority of the human genome is comprised of DNA from repetitive sequences and mobile genetic elements. Retrotransposons, mobile DNA that moves via an RNA intermediate, are the most abundant transposable elements and comprise approximately 40% of human genomic sequence. Of these retrotransposons, the non-long terminal repeat (non-LTR) long interspersed elements (LINE-1 or L1) retain a degree of autonomy, as some full-length (FL) L1s encode functional proteins necessary for retrotransposition [1]. Although over 99% of L1 sequences are inactive, either through deleterious mutations, 5′-end truncations, or internal rearrangements, bioinformatic and empirical analysis predict that 100 FL-L1s have the capacity for autonomous movement, and thus are termed retrotransposition-competent L1s (RC-L1s) [2]. The consensus RC-L1 is 6 kb and contains a 5′ untranslated region (5′ UTR) with an internal promoter, two nonoverlapping open reading frames (ORF1 and ORF2), and a 3′ UTR with its own polyadenylation signal. ORF1 encodes a 40 kd (p40) RNA binding protein that forms ribonucleoprotein particles with L1 RNA [3]. ORF2 encodes a 150 kd protein with an N-terminal endonuclease (EN) and a C-terminal reverse transcriptase (RT) domain [1].
Despite their small number, the 100 or so remaining RC-L1s continue to threaten the human genome. Recently, 82 FL-L1s with intact ORFs were cloned and their activity tested using a cell culture retrotransposition assay. Almost one-half (40/82) of the FL-L1s were shown to be retrotransposition competent, with a majority of the retrotransposition activity contributed by six “hot” L1s [2]. Although the potential for active L1s to greatly increase their copy number is limited by the propensity for truncations to occur at the 5′ end during integration, two highly active RC-L1s (L1RP and L1b-Thal) have been characterized that are the result of disease-causing, full-length de novo integration events [4]. Subsequent comparison with other RC-L1s showed that both L1RP and L1b-Thal exhibit high activity in cell culture and belong to a group of “hot” L1s responsible for most of the retrotransposition that occurs in our genome today [2]. In addition, characterization of cloned retrotransposition events using tagged-RC-L1 constructs in cultured cancer cells indicate that ∼ 10% of L1 insertions are accompanied by large chromosomal rearrangements, suggesting that active L1s could also lead to genomic instability [5, 6]. Furthermore, an increasing number of reports using advanced molecular techniques illustrate that L1s continue to negatively impact the fitness of the genome, either through de novo retrotransposition resulting in insertional mutagnesis, or as the result of unequal recombination between dispersed L1s and gene sequences [7–10].
Undoubtedly, both positive and negative factors continue to regulate L1 activity. For example, experiments using tagged retrotransposition-incompetent constructs (ie, Alu, pseudogene, and mutant L1s) demonstrate that nonautonomous RNAs are mobilized in trans by the L1 machinery at a much lower frequency compared to the RC-L1 that encoded them [11, 12]. This characteristic, known as cis-preference, limits the ability of nonautonomous retrotransposons to form functional RNPs, thereby preventing the accumulation of dead-end intermediates. In fact, cis-preference helps ensure the survival of the small number of RC-L1s that would otherwise compete with nonautonomous retrotransposons for limited host factors. The idea that RC-L1 might be under purifying selection, as well as various ways that L1s can negatively impact the genome, argues in favor of multiple mechanisms to regulate L1 activity. Considerable experimental evidence exists that RNA interference (RNAi) represses the activity of many different transposable elements in other eukaryotes, leading to speculation that RNAi might act in a similar manner against human L1s.
RNAi is a conserved eukaryotic mechanism in which double-stranded RNA (dsRNA) recognizes homologous mRNA transcripts and causes sequence-specific inhibition of gene expression through a number of mechanisms (Figure 1) [13]. RNAi is initiated by cleavage of endogenous long dsRNA or short-hairpin RNA (shRNA or pre-miRNA) precursors by the RNase III enzyme Dicer into 21–25 nucleotide small interfering RNA (siRNA) or microRNA (miRNA) effector molecules [14, 15]. The siRNAs, which are perfectly complementary to their target, recognize their cognate mRNA and become associated with a large multiprotein complex referred to as the RNA-induced silencing complex (RISC) that destroys target mRNAs by endonucleolytic cleavage at regions homologous to the siRNA [16, 17] (Figure 1(a)). miRNAs, on the other hand, are imperfectly matched with their target sequences and associate with homologous mRNAs in a ribonucleoprotein complex resulting in sequence-specific reduction of gene expression through translation inhibition [13, 18] (Figure 1(b)). In addition to gene silencing at the posttranscriptional level (ie, siRNA-mediated degradation or miRNA-mediated translation inhibition), siRNAs targeting promoter regions in genomic DNA can bring about DNA and histone methylation, resulting in promoter shutdown in a process termed transcriptional gene silencing (TGS) (Figure 1(c)) [13].
Figure 1.
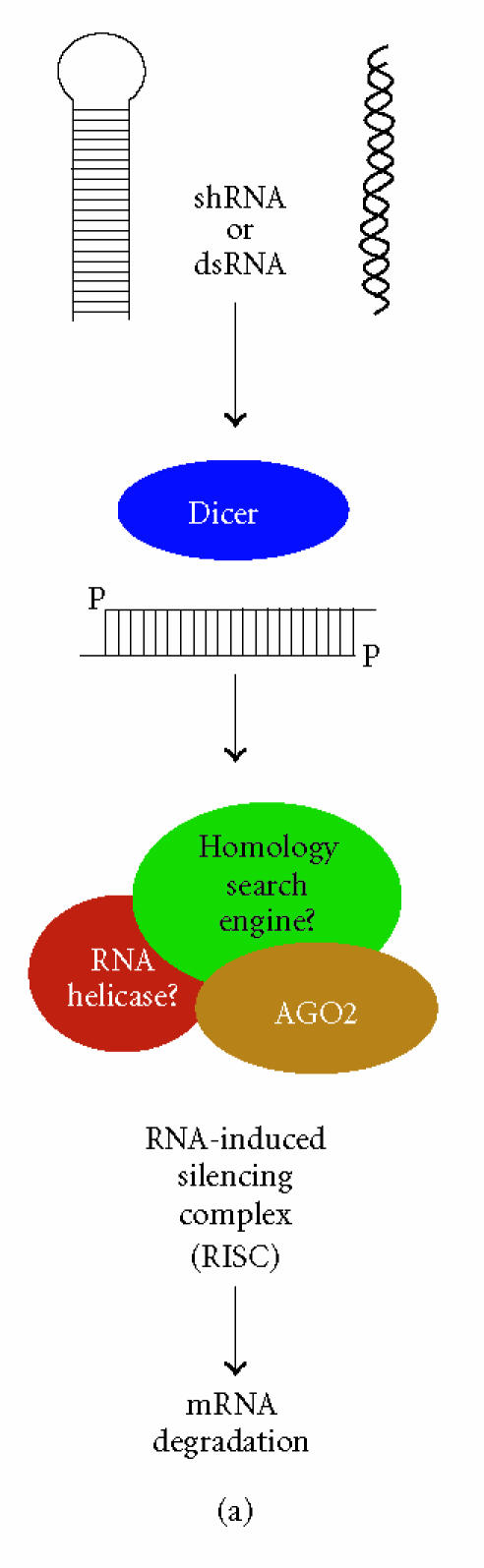
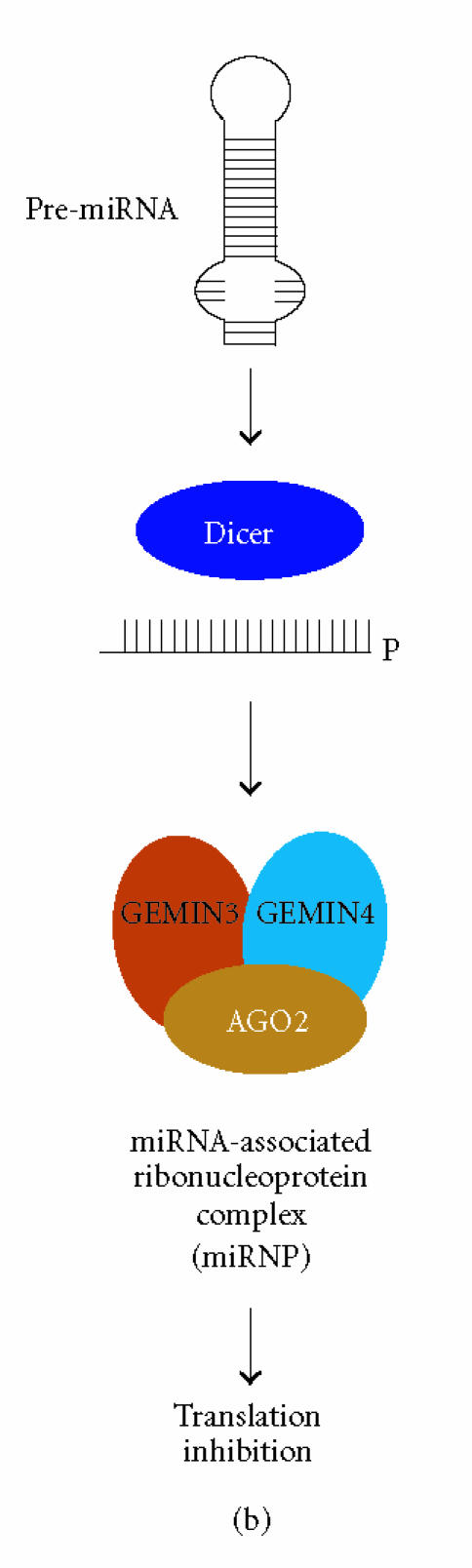
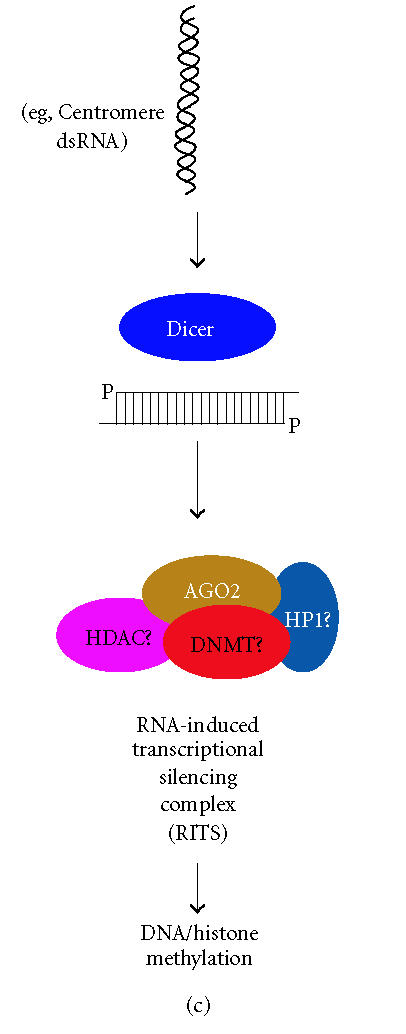
RNAi-based gene silencing pathways in H sapiens. (a) Dicer cleaves long double-stranded RNA (dsRNA) or short-hairpin RNA (shRNA) into functional siRNA with characteristic 3′ overhangs. siRNAs are incorporated into RISC, recognize the target mRNA through an unknown subunit(s), and cleavage is performed by AGO2. (b) Precursor microRNAs (pre-miRNA), which themselves are a cleavage product of a primary microRNA transcript, are further processed by Dicer into functional miRNAs that associate with AGO2 into a miRNA ribonucleoprotein (miRNP). miRNPs recognize their target mRNAs resulting in translation inhibition by an undefined mechanism. (c) Transcriptional gene silencing is initiated by Dicer-mediated cleavage of long dsRNA (eg, centromere dsRNA) into siRNA that associate with the RITS complex. Putative components of H sapiens RITS are depicted: AGO-argonaute; DNMT-DNA methyltransferase; HDAC-histone deacetylase; HP1-heterochromatin protein 1.
RNAi SUPPRESSES TRANSPOSABLE ELEMENTS IN MANY OF EUKARYOTES
The genetic link between RNAi and control of mobile genetic elements was initially established following EMS mutagenesis screens of Caenorhabditis elegans. Several C elegans mutants deficient in RNAi also show increased activity of DNA transposons, specifically Tc1, Tc3, and Tc5, as demonstrated by Southern blot analysis for Tc-directed insertions (Table 1) [19, 20]. Further screens in C elegans demonstrated that while not all genes necessary to RNAi are also required for transposon silencing, there is substantial cross-talk between the two regulatory pathways [21]. Additional evidence supporting a role for RNAi in silencing both transposons and retrotransposons has been demonstrated through genetic analysis in a number of other eukaryotes. One problem has been translating the results obtained in these model eukaryotes to the more complex human genome. Fortunately, the rich bioinformatics resources spawned from the genome sequencing efforts over the last decade permit the identification of human orthologs of essential RNAi components.
Table 1.
Eukaryotic RNAi orthologs involved in silencing transposable elements. H sapiens orthologs, if present in the Homologene database, are indicated. N.D. implies not determined.
| Organism | RNAi genes implicated in silencing TEs | Human ortholog | Transposable element silenced | siRNAs detected? | Reference(s) |
| Caenorhabditis elegans | mut-7 | — | Tc1, Tc3, Tc5 DNA transposons | Terminal inverted repeat (TIR) of Tc1 | [19–21, 48] |
| rde-2 | AGO2 | ||||
| mut-16 | — | ||||
| mut-14 | — | ||||
| Drosophila melanogaster | piwi | PIWI | Gypsy ERV | 5′ UTR | [32, 33] |
| Copia retrotransposon | N.D. | ||||
| Mdg1 retrotransposon | N.D. | ||||
| Trypanosma bruceii | Ago1 | AGO2 | Ingi retroposon | ORF 1 | [29, 30] |
| SLACS retrotransposon | ORF 1 and 3′ UTR | ||||
| Neurosporra crassa | qde-2 | AGO2 | Tad retrotransposon | ORF 1 and ORF 2 | [28] |
| dcl1/dcl2 | DICER1 | ||||
| Mus musculus | Dicer-1 | DICER-1 | LINE-1 | N.D. | [24, 25] |
| Intracisternal A particle | N.D. | ||||
| Arabidopsis thaliana | Ago4 | — | AtSN1 retroelement | AtSN1* | [31] |
*AtSN1 siRNA was determined by Northern blot with a full-length 159 nucleotides sense AtSN1 RNA probe.
Human cells encode one Dicer (DCR) protein, an enzyme with two RNase III domains that forms an intramolecular dimer to cleave dsRNA in a processive manner producing 21–25 nucleotide siRNAs [22]. The early embryonic lethality observed in mice with the Dicer null genotype (Dcr-1 −/−) confirms an essential role for Dicer in mammalian development. Unfortunately, the establishment of mouse embryonic fibroblast lines for further study has been hampered by the early death (E7.5) of Dicer null embryos [23]. To provide a more favorable system to study the role of Dicer in controlling mammalian retroelements, Dicer-deficient mouse embryonic stem (ES) cells were developed. Increased transcription of murine L1 elements was observed in the absence of Dicer, but not wild-type ES cells, providing the first direct evidence that RNAi controls the expression of murine L1 retrotransposons [24]. The observed increase in L1 expression was measured by quantitative RT-PCR using primers homologous to the murine L1 5′ UTR, presumably allowing quantification of transcripts originating from the ∼ 3000 RC-L1s that inhabit the C57/BL6 genome. In addition, transcripts from intracisternal A particles (IAPs), an active murine LTR-retrotransposon, were also elevated in the absence of Dicer. This report supports earlier work in which IAP and murine endogenous retrovirus-L transcripts were up-regulated following injection of anti-Dicer dsRNA into 2- and 8-cell stage mouse embryos [25]. As Dicer activity is necessary for limiting transcription of both non-LTR as well as LTR containing retrotransposons, one is not reaching to propose Dicer-mediated cleavage of endogenous retrotransposon-derived dsRNA into siRNA functions in human cells.
The siRNA produced by Dicer is handed off to the RNA-induced silencing complex (RISC). While the exact components of Homo sapiens RISC remain to be completely characterized, siRNA-mediated knockdown of in HeLa cells, as well as gene targeting experiments in mice, demonstrate that the RISC-component AGO2 is essential for target mRNA cleavage (Figure 1(a)) [26, 27]. Selective inactivation of AGO2 orthologs in lower eukaryotes demonstrates that RISC-associated Ago proteins are required for silencing both DNA transposons and retrotransposons. For example, loss of the AGO2 ortholog qde-2 in Neurospora crassa leads to increased expression of the LINE-like retrotransposon, Tad. Moreover, deletion of both Dicer genes causes an increase in Tad activity, linking the initiation step in RNAi to non-LTR retrotransposon silencing [28]. An interesting aspect of the analysis of Tad retrotransposition is that the Neurospora genome, which is devoid of active transposons through the action of efficient homology-dependent gene silencing mechanisms such as repeat-induced point mutations (RIP), requires an intact RNAi response to respond to the introduction by transformation of an active Tad element. Thus, perhaps one role of RNAi in higher eukaryotes is to permit a rapid and potent response to the sudden activation of retrotransposons.
In addition to LINE-like Tad retrotransposons, increased transcript levels of the Ingi and SLACS retroposon elements are observed in cells lacking Ago1, the AGO2 ortholog of Trypanosoma brucei RISC [29]. Several other spontaneous or induced AGO mutants, such as the Arabidopsis Ago4 and Drosophila piwi mutants, also show elevated levels of retrotransposons [30–33]. Thus, genetic evidence from a variety of organisms links both the initiation step (Dicer) and RISC-mediated effector step (AGO) of RNAi to the control mobile genetic elements. Moreover, the fact that different classes of transposable elements (DNA transposons, LTR and non-LTR retrotransposons, and endogenous retrovirus sequences) are up-regulated in the absence of the RNAi machinery supports the generalization that RNAi is part of the eukaryotic innate immune system to protect the genome from the mutational load of parasitic sequences [34] (Table 1).
DOES RNAi CONTROL LINE-1 ACTIVITY IN HUMAN CELLS?
So far, there is no direct evidence that the RNAi pathway in human cells protects the genome from the activity of L1s. Direct genetic evidence has been hard to come by in human cells because of the difficulty in inhibiting RNAi gene function. For other model eukaryotes such as C elegans and Schizosaccharomyces pombe, the high rate of homologous recombination (HR) and ability to perform large-scale genetic screens, permits the study of mutant phenotypes through insertion and/or inactivation of specific genes [35]. Moreover, the recent application of RNAi technology to selectively inhibit gene function in mammalian cells both in culture and in vivo had made it less necessary to rigorously pursue methods that enhance the efficiency of HR in mammalian cells. Although several genetic screens in mammalian cells have been conducted using shRNA libraries, one can appreciate this RNAi-mediated approach would be problematic for studying the role that RNAi plays in controlling human L1s [36, 37]. It is possible to achieve transient inhibition of the RNAi pathway by transfecting human cells with large quantities (> 50 nM) of siRNA targeting one of the RNAi components (eg, DICER or AGO2) [15, 38]. However, functional inhibition of the RNAi pathway is directly proportional to transfection efficiency and varies between cell lines (unpublished observations). In addition, some virus products are inhibitors of RNAi, either by successfully competing with endogenous dsRNA for Dicer, as is the case for the adenovirus VA1 noncoding RNA, or by sequestering siRNA in an inactive complex [39–41]. Although one group reported efficient down-regulation of Dicer in HeLa cells using a trans-cleaving hammerhead ribozyme, only transient knockdown of Dicer expression was achieved and they did not demonstrate functional inhibition of the RNAi pathway [42].
In the absence of data showing increased L1 activity in cells with an impaired RNAi pathway, the detection and cloning of L1-derived siRNAs would support a role for RNAi in controlling L1s. Efforts to clone the small RNA fraction from HeLa cells failed to find microRNAs (miRNAs) produced from LINE-1, suggesting that if endogenous L1 miRNAs are produced, they are present at low levels or in specific cell types [43]. This initial cloning effort relied on high throughput sequencing after annealing linker molecules to the small RNA fraction purified from HeLa cells and might overlook miRNAs from repetitive elements. Indeed, endogenous siRNAs homologous to centromere repeats were not cloned using this approach, despite being detected by RNase protection and Northern blot analyses in chicken DT40 and murine ES cells, respectively [24, 44]. Restriction of L1 siRNAs specific cell types, such as primordial germ tissue and/or gametes, would explain why earlier characterization of endogenous siRNAs in human cervical carcinoma cells failed to detect L1 siRNAs. Since L1s that retrotranspose in gametes insure passage of their genetic information to the next generation without impacting host fitness through somatic mutagenesis, the cell might combat this threat by producing L1 siRNA at a specific time during gametogenesis. Despite the advantage for L1s to restrict their expression in germ cells, immunohistochemical analysis detected L1 ORF translation products (ORF1p and ORF2p) in adult and fetal testicular tissue, as well as Sertoli, Leydig, and vascular endothelial cells [45, 46]. Furthermore, a single case of insertional mutagenesis by L1 in somatic tissue has been reported [47]. Consequently, the threat posed by RC-L1s and functional ORF proteins is not limited to the germline, and L1 siRNAs might also be present in somatic cells. Moreover, as the amount of FL-L1 RNA in cultured somatic cells is relatively low compared to L1 expression from established germ cell tumors, somatic cells seem a fitting place for posttranscriptional degradation of FL-L1 RNA by siRNA to occur.
In lower eukaryotes where classical genetics has established a direct link between RNAi and the control of mobile genetic elements, siRNAs have been detected for both transposons and retrotransposons. For example, siRNAs derived from the LINE-like Tad retrotransposon were detected by Northern blotting of total RNA from Neurospora crassa qde-2 mutants, but not wild-type progeny [28] (Table 1). qde-2 is the AGO2 ortholog of N crassa RISC, and qde-2 mutants are viable, but defective in RNAi. Tad-specific siRNAs were detected with probes homologous to the Tad ORF1 or ORF2, indicating that siRNAs were produced along the length of the element. In C elegans, RNase protection analysis successfully detected Tc1 dsRNA produced by read-through transcription of endogenous promoters, as well as Tc1 siRNA in the germ line of wild-type and RNAi-deficient worms (Table 1). In contrast to Tad siRNAs, endogenous siRNAs from the C elegans DNA transposons were not derived from the transposase ORF, but were detected with probes complementary to the inverted repeats [48]. The fact that C elegans mutator strains also show increased mobility of other DNA transposons such as Tc3 and Tc5, suggests that the C elegans RNAi is not specific to one element and RNAi might be a general defense mechanism against transposon activity. Endogenous siRNAs homologous to retrotransposons have also been detected by Northern blot in Arabidopsis thaliana and Drosophila melanogaster (Table 1) [31, 32].
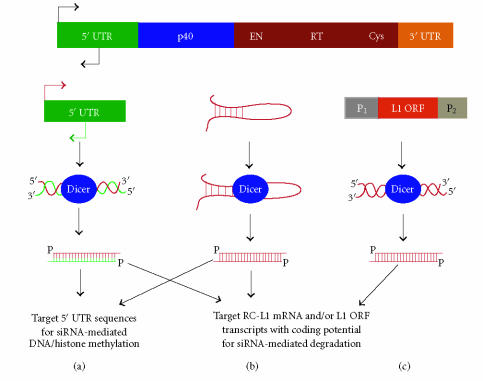
One requirement for the production of L1 siRNA would be transcription of antisense L1 RNA that could hybridize with L1 sense RNA to form dsRNA followed by Dicer-mediated processing into siRNAs. An early study of L1 expression demonstrated that large quantities of both sense and antisense L1 RNA of variable size greater than 1 kb are present in total RNA of a human teratocarcinoma cell line, but not in the cytoplasmic RNA fractions where Dicer processing of L1 dsRNA might occur [49]. The expression profile of L1 sequences is particularly complicated, not only because the ∼ 3000 FL-L1s that reside in the human genome contain an internal Pol II promoter that could remain transcriptionally active, but strong cellular promoters nearby presumably inactive L1s could result in the expression and translation of unwanted L1 ORF products [50] (Figure 2(c)). Therefore, the production of L1 dsRNA and its conversion by Dicer into L1 siRNA might simply be a consequence of the large number (> 500 000 copies/diploid genome) of L1 sequences and their proximity to transcriptionally active, endogenous promoters (Figure 2(c)). The activity of adjacent promoters also establishes the possibility that L1 dsRNA or siRNA could form through simple diffusion of complementary L1 transcripts expressed from distant loci. In addition to the activity of cellular promoters, regions of the L1 mRNA that form stable hairpin structures greater than 21 nucleotides might also be subject to Dicer processing into siRNA (Figure 2(b)). To date, no L1 hairpin structures have been defined biochemically, although recombinant human Dicer efficiently converts in vitro transcribed L1 dsRNA into functional siRNA [51].
Figure 2.
Proposed ways RNAi can control L1 activity. The consensus RC-L1 is depicted above. (a) L1 dsRNA produced from the 5′ UTR sense and antisense promoters is processed by Dicer and can target transcripts originating from RC-L1s for degradation. Alternatively, this siRNA can also initiate histone and DNA methylation resulting in silencing of the L1s promoter. (b) Regions of L1 mRNA that form stable hairpins through intramolecular base pairing could be Dicer substrates. The resulting siRNA is capable of a number of responses. (c) L1 dsRNA produced by read-through transcription from opposing cellular promoters is converted into siRNA that can target RC-L1 or ORF transcripts for degradation.
Instead of relying on adjacent promoters for transcription, the production of sense/antisense L1 dsRNA might take advantage of a unique feature of the L1 5′ UTR; the existence of an internal promoter that transcribes L1 sense RNA and an antisense promoter (ASP) within nucleotides +400 to +600 (with respect to the 5′-end of the L1) of the 5′ UTR that transcribes minus-strand L1 sequence in the opposite direction (Figure 2(a)) [52, 53]. In cell lines where the 5′ UTR sense promoter shows transcriptional activity, the L1 ASP is also active, albeit at lower levels [52, 54]. The resulting minus strand L1 RNA could anneal with plus strand L1 RNA originating from the same L1 5′ UTR region, or anneal with another 5′ UTR sense RNA by diffusion. Dicer could then convert the dsRNA derived from the L1's 5′ UTR into siRNA. It is important to recognize that 5′ UTR siRNA can act on transcripts arising from the L1s sense promoter as well as the L1s ASP (Figure 2(a)). As the mechanism for choosing which strand of the siRNA (sense strand targeting antisense message or antisense strand targeting sense message) is incorporated into RISC along with the target is not well understood, it is possible that siRNA produced from this unique region of the L1s 5′ UTR could generate two different RNAi responses [55, 56]. First, L1 retrotransposition could be kept in check by the antisense siRNA strand recognizing transcripts originating from RC-L1s. Additionally, the sense siRNA strand could target transcripts from the L1s ASP, thereby regulating the expression of certain endogenous genes through the action of a single pool of L1 5′ UTR siRNAs [53].
As of yet, short duplex RNAs derived from L1s await characterization, possibly owing to low-level expression in specific cell types. Solution hybridization using radiolabelled RNA probes from conserved regions of the L1s 5′ UTR offers a sensitive method to detect endogenous 5′ UTR siRNAs. For the detection of L1 siRNA, it will be necessary to distinguish short, single-stranded L1 RNA that might hybridize to the riboprobe and be mistakenly detected as L1 siRNA, from the real L1 siRNA duplexes, which being double-stranded are resistant to RNase A activity in the presence of high salt [48]. A further issue complicating the detection of L1 siRNAs by ribonuclease digestion is the fact that single nucleotide mismatches between endogenous L1 siRNAs and the riboprobe might cause cleavage and detection of protected fragments that are smaller than the predicted 21–25 nucleotides size for siRNA. Careful design of 5′ UTR riboprobes should limit potential problems caused by single nucleotide mismatches. For example, one could restrict detection of siRNAs to a specific L1 subfamily, such as Ta-1d, which harbors a deletion at position 72 of the 5′ UTR and distinguishes this youngest L1 subset from the slightly more divergent Ta-1nd [57].
CONCLUSION
There is ample experimental evidence, through genetic manipulation and biochemical analysis, that RNA interference controls the activity of transposable elements in a variety of eukaryotes such as A thaliana, S pombe, C elegans, and M musculus [19–21, 24, 25, 28–33]. In addition, since the RNAi response can efficiently limit retrotransposition of an RC-L1 when introduced into transformed human cells, there are no barriers per se to siRNA-mediated degradation of L1s. The inability to uncover direct evidence that RNAi may control L1 activity is not due to a lack of effort, as several groups are pursuing experiments to assess the interaction between RNAi and human L1s. The difficulty in studying the activity of endogenous human L1s in cells with an impaired RNAi pathway has slowed progress in showing a role for RNAi in suppressing L1s. As current Dicer- and Ago2-null mice show early embryonic lethality, the use of conditional gene targeting through Cre-mediated excision of floxed-RNAi alleles will permit further assessment for the role of RNAi in L1 retrotransposition [23, 27]. Conditional gene targeting and deletion of Dicer in the T cells causes loss of microRNA processing linked to impaired T cell differentiation [58]. These Dicer-deficient T cells are viable, but lack Dicer activity, thus providing a distinct Dicer-null population for which retrotransposon activity can be assessed. It is just a matter of time before proper experiments, combined with dogged determination, provide direct evidence that human L1s are, to some degree, constrained by the RNAi pathway.
ACKNOWLEDGMENTS
I would like to thank Lars Aagard, Kevin Morris, and John Rossi for critical review of this manuscript and helpful comments. The author is supported by the Beckman Fellowship from the Arnold and Mabel Beckman Foundation.
References
- 1.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annual Review of Genetics. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 2.Brouha B, Schustak J, Badge RM, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(9):5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Human Molecular Genetics. 2005;14(21):3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 4.Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Human Molecular Genetics. 1999;8(8):1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 6.Symer DE, Connelly C, Szak ST, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 7.Burwinkel B, Kilimann MW. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. Journal of Molecular Biology. 1998;277(3):513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 8.Kumatori A, Faizunnessa NN, Suzuki S, Moriuchi T, Kurozumi H, Nakamura M. Nonhomologous recombination between the cytochrome b558 heavy chain gene (CYBB) and LINE-1 causes an X-linked chronic granulomatous disease. Genomics. 1998;53(2):123–128. doi: 10.1006/geno.1998.5510. [DOI] [PubMed] [Google Scholar]
- 9.Suminaga R, Takeshima Y, Yasuda K, Shiga N, Nakamura H, Matsuo M. Non-homologous recombination between Alu and LINE-1 repeats caused a 430-kb deletion in the dystrophin gene: a novel source of genomic instability. Journal of Human Genetics. 2000;45(6):331–336. doi: 10.1007/s100380070003. [DOI] [PubMed] [Google Scholar]
- 10.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nature Genetics. 1998;19(1):19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 11.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genetics. 2000;24(4):363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 12.Wei W, Gilbert N, Ooi SL, et al. Human L1 retrotransposition: cis preference versus trans complementation. Molecular and Cellular Biology. 2001;21(4):1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meister G, Tuschl T. Mechanisms of gene silencing by doublestranded RNA. Nature. 2004;431(7006):343–349 . doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 14.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Jeon K, Lee J-T, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. The EMBO Journal. 2002;21(17):4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293(5532):1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Current Biology. 2004;14(9):787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Mourelatos Z, Dostie J, Paushkin S, et al. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes & Development. 2002;16(6):720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabara H, Sarkissian M, Kelly WG, et al. The rde-1 gene, RNA interference, and transposon silencing in C elegans . Cell. 1999;99(2):123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 20.Ketting RF, Haverkamp THA, van Luenen HGAM, Plasterk RHA. Mut-7 of C elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99(2):133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 21.Vastenhouw NL, Fischer SEJ, Robert VJP, et al. A genome-wide screen identifies 27 genes involved in transposon silencing in C elegans. Current Biology. 2003;13(15):1311–1316. doi: 10.1016/s0960-9822(03)00539-6. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nature Genetics. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 24.Kanellopoulou C, Muljo SA, Kung AL, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Development. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Developmental Biology. 2004;269(1):276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 26.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Molecular Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 28.Nolan T, Braccini L, Azzalin G, De Toni A, Macino G, Cogoni C. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa . Nucleic Acids Research. 2005;33(5):1564–1573. doi: 10.1093/nar/gki300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H, Djikeng A, Tschudi C, Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Molecular and Cellular Biology. 2004;24(1):420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djikeng A, Shi H, Tschudi C, Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24-26-nucleotide RNAs. RNA. 2001;7(11):1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 31.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299(5607):716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 32.Sarot E, Payen-Groschêne G, Bucheton A, Pélisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166(3):1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Research. 2005;33(6):2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vastenhouw NL, Plasterk RHA. RNAi protects the Caenorhabditis elegans germline against transposition. Trends in Genetics. 2004;20(7):314–319. doi: 10.1016/j.tig.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Hudson DF, Morrison C, Ruchaud S, Earnshaw WC. Reverse genetics of essential genes in tissue-culture cells: ‘dead cells talking’. Trends in Cell Biology. 2002;12(6):281–287. doi: 10.1016/s0962-8924(02)02281-x. [DOI] [PubMed] [Google Scholar]
- 36.Paddison PJ, Silva JM, Conklin DS, et al. A resource for largescale RNA-interference-based screens in mammals. Nature. 2004;428(6981):427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 37.Berns K, Hijmans EM, Mullenders J, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428(6981):431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 38.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 39.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426(6968):874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. Journal of Virology. 2004;78(23):12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. Journal of General Virology. 2004;85(pt 4):983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- 42.Kawasaki H, Taira K. Short hairpin type of dsRNAs that are controlled by tRNAVal promoter significantly induce RNAimediated gene silencing in the cytoplasm of human cells. Nucleic Acids Research. 2003;31(2):700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 44.Fukagawa T, Nogami M, Yoshikawa M, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nature Cell Biology. 2004;6(8):784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 45.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Molecular and Cellular Biology. 1994;14(4):2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ergün S, Buschmann C, Heukeshoven J, et al. Cell typespecific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. The Journal of Biological Chemistry. 2004;279(26):27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- 47.Miki Y, Nishisho I, Horii A, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Research. 1992;52(3):643–645. [PubMed] [Google Scholar]
- 48.Sijen T, Plasterk RHA. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426(6964):310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 49.Skowronski J, Singer MF. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(18):6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Molecular and Cellular Biology. 1990;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soifer HS, Zaragoza A, Peyvan M, Behlke MA, Rossi JJ. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Research. 2005;33(3):846–856. doi: 10.1093/nar/gki223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Molecular and Cellular Biology. 2001;21(6):1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nigumann P, Redik K, Mätlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79(5):628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- 54.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Research. 2003;31(16):4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 56.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 57.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Molecular Biology and Evolution. 2000;17(6):915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 58.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. The Journal of Experimental Medicine. 2005;202(2):261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]