Figure 1.
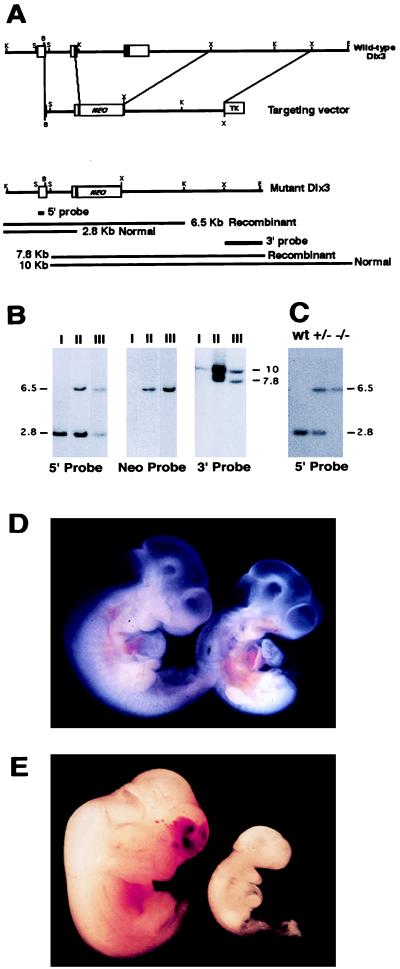
Targeted disruption of Dlx3. (A) Restriction map of the Dlx3 gene, the targeting vector, and the predicted restriction map after the homologous recombination event. The probes used and the predicted length of restriction fragments in the Southern blot analysis are shown. B, BamHI; K, KpnI; E, EcoRI; S, SmaI; X, XbaI; NEO, neomycin phosphotransferase gene; TK, thymidine kinase gene. (B) Genomic analysis of wild-type-derived ES cell DNA (lanes I) and two independently targeted ES cell clones (lanes II and III). Recombination was detected at the 5′ end by digesting ES cell DNA with KpnI/Asp718 and hybridizing the Southern blot with 5′ probe, and at the 3′ end by digesting DNA with EcoRI and SmaI and hybridizing with 3′ probe. A single integration event was confirmed by hybridizing KpnI/Asp718-digested DNA with probe Neo (Center). Numbers at sides of the figure indicate fragment size in kb. (C) Genotyping of tail-derived DNA revealing the expected pattern for wild-type (wt), heterozygous (+/−), and homozygous Dlx3-null (−/−) embryonic day 9.5 (E9.5) embryos. (D) Phenotype of wild-type (left) and Dlx3-null (right) E9.5 embryos genotyped by Southern blotting. (E) Phenotype of wild-type (left) and regressed Dlx3 −/− (right) E12.5 embryos genotyped by Southern blotting.