Figure 1.
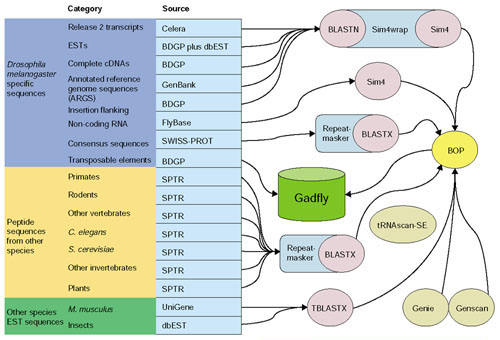
Gadfly data sources and analyses. This figure provides an overview of the pipeline analyses that flow into the central annotation database (Gadfly) and are provided to the curators for annotation. The D. melanogaster-specific datasets (dark blue) are one of the following: nucleic acids, peptides (from SPTR: SWISS-PROT/TrEMBL/TrEMBLNEW [31]), or transposable elements (the source of the sequences are listed in the light-blue column). The nucleic acids are aligned using sim4 and the peptides using BLASTX. The transposable elements are the product of a more detailed analysis [46] and their coordinates were recorded directly in Gadfly. The peptide datasets from other species (yellow) were obtained from SWISS-PROT and aligned using BLASTX. We used TBLASTX to translate (in all six frames) and align the rodent UniGene [47] and insect ESTs from dbEST [48] (green). For ab initio predictions on the genomic sequence we used Genie [42], Genscan [43] and tRNAscan-SE [44]. BOP was used to filter BLAST and sim4 results and parse all the results to output GAME XML; the results were recorded in Gadfly by loading the XML into the database.
