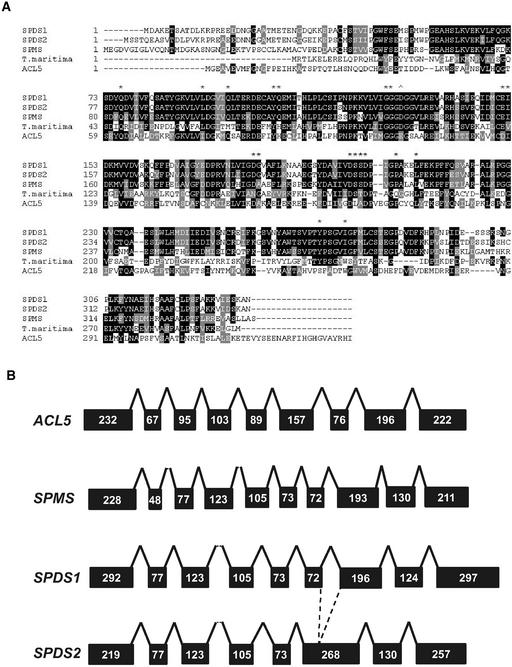
Figure 1.
Amino Acid Sequence Comparison and Genomic Organization of Arabidopsis Aminopropyl Transferase Proteins.
(A) Comparison of amino acid sequences of Arabidopsis and T. maritima SPDS proteins. The SPDS protein sequences were aligned and compared by Baylor College of Medicine Search Launcher (http://searchlauncher.bcm.tmc.edu). Black shading indicates identical amino acid residues, whereas similarities are highlighted in gray. Asterisks indicate the positions of residues that interact with the artificial inhibitor 5-adenosyl-1,8-diamino-3-thiooctane. The arrowhead indicates a residue of putrescine N-methyltransferase that deviates from the conserved residues of SPDS.
(B) Scheme of the intron/exon structure of the genomic regions of the SPDS1, SPDS2, SPMS, and ACL5 genes. Black boxes and solid lines represent exons and introns, respectively. Numbers refer to the nucleotides in each exon.