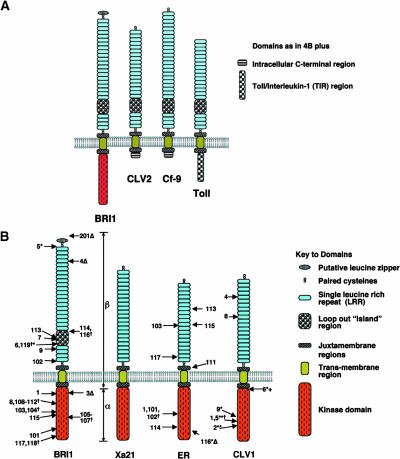
Figure 4.
Schematic Structure of BRI1 and Similar LRR-RLKs and LRR receptor–like Proteins.
(A) Similarities in LRR-domain topologies highlighting a similar location of loop-out island region (for details see main text). Source of primary sequence information: BRI1, BL signaling (Li and Chory, 1997); CLV2, LRR-R involved in control of plant meristem size and differentiation (Jeong et al., 1999); Cf-9, involved in plant pathogen signaling (Jones et al., 1994); and Toll, involved in Drosophila embryo development and immunity pathways (Hashimoto et al., 1988). Note: domains are not to scale and the N-terminal signal peptides are not shown.
(B) Domain topology of selected LRR-RLKs highlighting the location of sequenced mutations. BRI1, location of brassinolide insensitive 1 (Li and Chory, 1997) and additional alleles (Noguchi et al., 1999; Friedrichsen et al., 2000; Bouquin et al., 2001); Xa21, (Song et al., 1995); ER, erecta-1 (Torii et al., 1996) and additional alleles (Lease et al., 2001); CLV1, clavata-1 alleles (Clark et al., 1997). Missense mutations indicated by arrows are shown on the left, whereas nonsense, deletion (Δ) and insertion (+) mutations are indicated on the right. (†), independent mutations at the same position. All alleles are strong (null alleles) unless indicated. (*), weak alleles; (**), intermediate alleles. α and β indicate the approximate regions used in domain swaps between BRI1 and Xa21 (He et al., 2000). Note: domains are not to scale, and the N-terminal signal peptide is not shown. Also note: allele bri-201 is a deletion mutant in the N-terminal region that is not shown.