Figure 2.
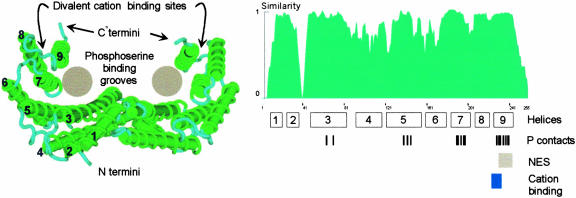
Model of the 14-3-3 Structure.
The structure of the main body of the protein is known to be a W-shaped clamp formed by two monomers, each of which is capable of binding phosphorylated peptides. The structure and positions of the extreme N and C termini are unresolved in the crystals, and the C termini likely form a movable flap that can seal the top of the clamp, perhaps depending on divalent cation binding. The conservation of the 14-3-3 protein sequences is evident when the similarity index among all Arabidopsis isoforms is plotted along the length of the molecule. Only three areas of the 14-3-3s show low degrees of similarity: the N terminus, the C terminus, and the region between helices 2 and 3. Shown below the similarity profile are the locations of the helices in the crystal structure as well as the locations of the residues responsible for contacting the phosphopeptide (P contacts) (Rittinger et al., 1999), the nuclear exclusion sequence (NES) (Rittinger et al., 1999), and the cation binding site (Lu et al., 1994). The crystal structure was taken from the protein database 1QJB and was modeled using Cn3D. The similarity profile among Arabidopsis 14-3-3 was calculated using the algorithms available in Vector NTI 7.0 (InforMax, Bethesda, MD).
