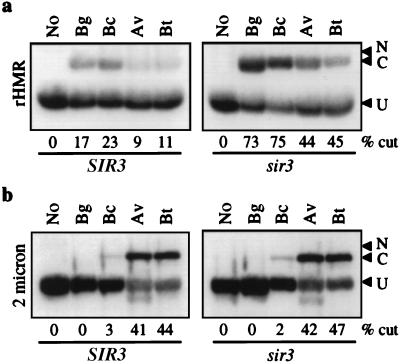
Figure 3.
Restriction endonuclease accessibility of partially purified chromatin rings. Samples were prepared from strain YCL2, which was transformed with pRS415-RecR and either pSIR3-HA or pRS416 to yield SIR3 and sir3 strains, respectively. Chromatin rings were isolated in RI buffer containing 150 mM KCl and 10% glycerol. After digestion and electrophoresis, blots were hybridized with either a probe to detect the rHMR ring (a) or a probe to detect the entire 2-μm plasmid (b). Linearized, closed-circular, and nicked species are denoted as C (cut), U (uncut), and N (nicked), respectively. Enzymes corresponding to unique sites in rHMR were used as indicated above each lane (see Fig. 1 for map and legend). In lane 1, no enzyme (No) was used. The extent of enzymatic digestion is reported as the percentage of band C relative to the sum of bands C and U in each lane. The 2-μm plasmid contains no BglII sites and one BclI site. There are multiple sites for AvaII and BstBI yet singly cut species are observed because of partial cleavage of the chromatin templates.