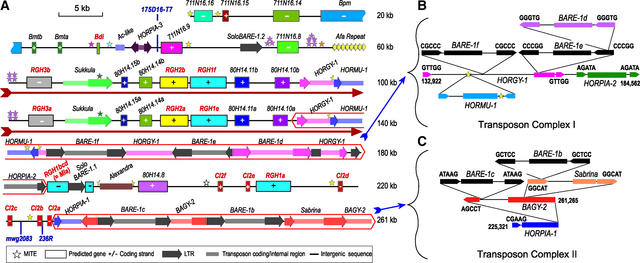
Figure 1.
Sequence Annotation of the Barley Mla Locus from cv Morex.
BAC 711N16 (top; 112,178 nucleotides) was sequenced at ×12 redundancy, and BAC 80H14 (bottom; 158,773 nucleotides) was sequenced at ×15 redundancy; sequence error is <1 bp/10 kb. The two BACs are joined by 9686 nucleotides of overlapping sequence encompassing RGH3a and the 5′ end of the Sukkula retrotransposon. The GC content of 261,265 nucleotides of finished sequence is 44.9%, the CpG ratio is 4.2%, the observed/expected CpG ratio is 0.83, and the CpG/GpC ratio is 1.21. These values are consistent with barley genomic contigs encompassing barley Mlo (Panstruga et al., 1998) and Rar1 (Shirasu et al., 2000). + indicates forward strand transcription, and − designates complementary strand transcription.
(A) Annotation of the 261,265-nucleotide region. The dark-red text indicates genes associated with defense responses, and the dark-red arrow is below the 40-kb tandem duplication. Different colors show different families of genes or transposable elements.
(B) Nested transposon complex I on BAC 80H14 between 132,922 and 184,562 nucleotides.
(C) Nested transposon complex II on BAC 80H14 between 225,321 and 261,265 nucleotides.
Retrotransposon placement was determined by the 5-bp LTR inverted repeat insertion signature (6-bp direct repeat shown in letters) at the two ends of each element. This direct repeat marks the boundary for each retroelement.