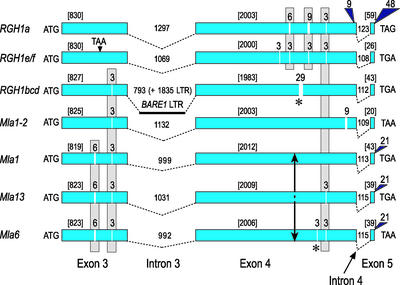
Figure 2.
Structural Comparison of Morex RGH1 Paralogs with Functional Mla Specificities from Accessions C.I. 16137 (Mla1), C.I. 16151 (Mla6), and C.I. 16155 (Mla13).
Deletions in the coding regions are designated by vertical white bars, and insertions are designated by navy blue triangles. Above each vertical white bar or blue triangle is the number of nucleotides for each InDel. InDels that delete an amino acid in the solvent-exposed region are designated with asterisks below the InDel. The number of nucleotides in each exon are shown in brackets. The number of nucleotides in each intron are specified above the intron. The divergence point in the predicted LRR domain of MLA1/MLA6 is marked by an arrow. The BARE-1 LTR in the major intron of RGH1bcd is designated by a solid horizontal line. RGH1e and 1f are both within the 40-kb duplication and differ by one nucleotide but no amino acid change.