Figure 9.
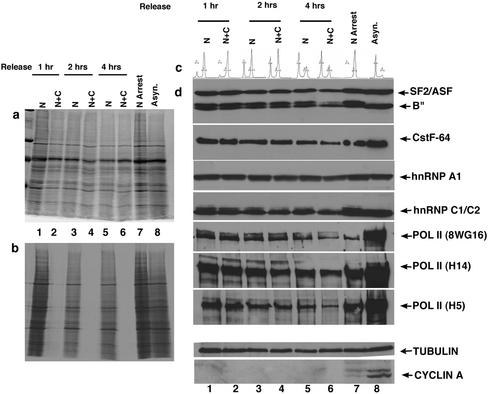
RNA processing factors exhibit a low turnover rate during mitosis. Cells blocked in prometaphase by nocodazole treatment were incubated with (N + C) or without (N) cycloheximide and pulsed with [35S]methionine during release from the mitotic block. Aliquots of cells were collected at indicated times and efficacy of the protein synthesis block was analyzed by measuring incorporated [35S]methionine by autoradiography (b) (see MATERIALS AND METHODS). The corresponding Coomassie-stained gel is shown in a. Lanes 1, 3, and 5 are cells treated with nocodazole for 14–18 h and collected at 1, 2, and 4 h postrelease, respectively. Lanes 2, 4, and 6 represent cells treated with nocodazole and cycloheximide at indicated time points. Lanes 7 and 8 represents nocodazole-arrested cells and an asynchronous population, respectively. DNA content of the respective cell populations indicated in d, 1–8, was determined by FACS analysis and is shown in c. Cells treated with or without cycloheximide were collected at indicated time points and extracted for immunoblot analysis of components of the gene expression machinery (d). The results show a low turnover of splicing factors (SF2/ASF and B"), CstF-64, hnRNP A1, hnRNPC1/C2, and various forms of RNA pol II during mitosis. Tubulin and cyclin A are shown as internal controls for protein loading and cell cycle progression, respectively.