Table 1.
Time-resolved fluorescence data and analysis
| Sample | τdecay,* ns |
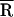 (Å)† (Å)†
|
σ (Å)† |
|---|---|---|---|
| No heat or NCp15 | |||
| tRNA-FL | 4.3 | — | — |
| TMR–tRNA–FL | 0.3 (85)‡ | 32 | 6 |
| tRNA–FL/T | 4.2 | — | — |
| TMR–tRNA–FL/T | 0.4 (74)‡ | 34 | 7 |
| Heat-annealed | |||
| tRNA–FL/T | 4.3 (76)§ | — | — |
| TMR–tRNA–FL/T | 3.9 (70)§ | 73 | — |
| NCp15-annealed | |||
| tRNA–FL | 4.3 (68)¶ | — | — |
| TMR–tRNA–FL | 0.4 (77)‡ | 35 | 5 |
| tRNA–FL/T | 4.3 (68)§ | — | — |
| TMR–tRNA–FL/T | 3.9 (52)§ | 73 | — |
Values are averages of at least five determinations. Numbers in parentheses represent typical amplitudes (percent total signal) with SD ≤ 8%.
Based on a Gaussian distribution [g(R)] analysis, where 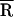 is the mean donor–acceptor distance and σ is the SD of g(R). SD ≤ 3 Å for
is the mean donor–acceptor distance and σ is the SD of g(R). SD ≤ 3 Å for 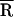 and ≤2 Å for σ.
and ≤2 Å for σ.
Mean donor lifetimes (SD ≤ 36%) from the Gaussian distribution analysis. A minor component (τD ∼ 4.3 ns), which may be the result of FL’s inability to undergo efficient FRET because of misfolded tRNA, was subtracted from the raw data before fitting was performed.
Based on a bi-exponential fit (SD ≤ 5%). A second component (τ ∼ 1.2 ns) can be resolved from these data.
Based on a single measurement.
