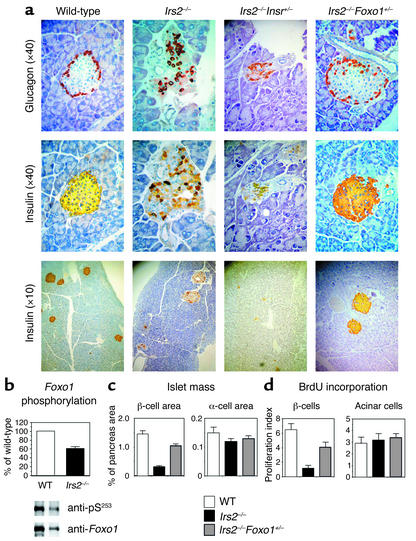
Figure 2.
(a) Pancreatic histology in mice with targeted null alleles of Irs2 and Foxo1. We stained pancreatic sections from 8-week-old mice of the indicated genotypes with anti-insulin and anti-glucagon antibodies. (b) Foxo1 phosphorylation in Irs2–/– islets. We transduced islets from wild-type and Irs2–/– mice with adenovirus encoding hemagglutinin-tagged wild-type Foxo1. Following immunoprecipitation with anti-hemagglutinin antibody, we carried out immunoblotting with anti–phospho-Foxo1S253. We then stripped and reprobed the blots with anti-Foxo1 antibody. One of three experiments is shown, and mean phosphorylation ± SEM is summarized in the graph above (WT, white bar; Irs2–/–, black bar), following scanning densitometry and normalization for total Foxo1 protein levels. (c) Islet morphometry. We quantitated β and α cell area in mice of the indicated genotypes using NIH Image 1.60 analysis software. Results are expressed as the percentage of total surveyed area containing insulin- or glucagon-immunoreactive cells. (d) Measurements of mitotic indices in pancreatic cells. We labeled cells traversing S phase of the replication cycle with BrdU in vivo and visualized them in pancreatic sections using double immunohistochemistry with anti-BrdU and anti-insulin antibodies. We calculated labeling indices by counting BrdU-positive cells as percentage of total number of cells within each microscopic field. We scored exocrine acinar cells based on their morphological appearance. Results represent the mean ± SEM of 12 sections from at least four mice for each genotype. For each genotype, we scored at least 50 microscopic fields.