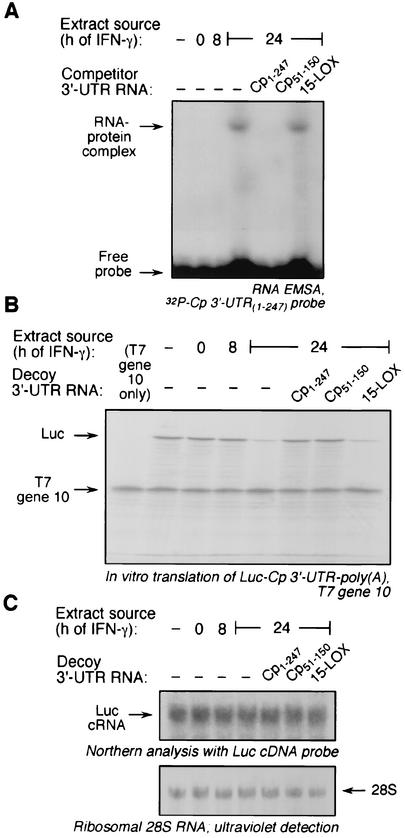
FIG. 2.
Protein binding to Cp 3′-UTR(51-150). U937 cells were incubated with IFN-γ (500 U/ml) for 0, 8, or 24 h, and cytosolic extracts were prepared. (A) Protein binding to Cp 3′-UTR(51-150) by competitive EMSA. Cell extracts were incubated with 32P-labeled, full-length Cp 3′-UTR. In competition experiments, the extracts were first incubated with a 10-fold molar excess of unlabeled, full-length Cp 3′-UTR (Cp1-247), Cp 3′-UTR(51-150) (Cp51-150), or the 15-LOX 3′-UTR (15-LOX) before addition of the labeled probe. RNA-protein complexes were resolved by electrophoresis on a nondenaturing, 5% polyacrylamide gel and detected by autoradiography. (B) Binding activity of Cp 3′-UTR(51-150) used as a decoy to overcome translational silencing. Capped, Luc-Cp 3′-UTR-poly(A) cRNA was subjected to in vitro translation in a rabbit reticulocyte lysate containing [35S]methionine and cytosolic extracts from U937 cells treated with IFN-γ (500 U/ml) for 8 or 24 h. A 50-fold molar excess of Cp1-247, Cp51-150, or 15-LOX 3′-UTR was added as decoy. Capped, T7 gene 10 mRNA was addedto each lysate as a loading and specificity control. Newly translated, 35S-labeled Luc and T7 gene 10 were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and detected by fluorography. (C) RNA from the reticulocyte lysates was isolated and subjected to Northern analysis using a radiolabeled, Luc cDNA probe (upper panel). rRNA from reticulocyte lysate was detected by UV and used as a loading control (lower panel, image inverted).