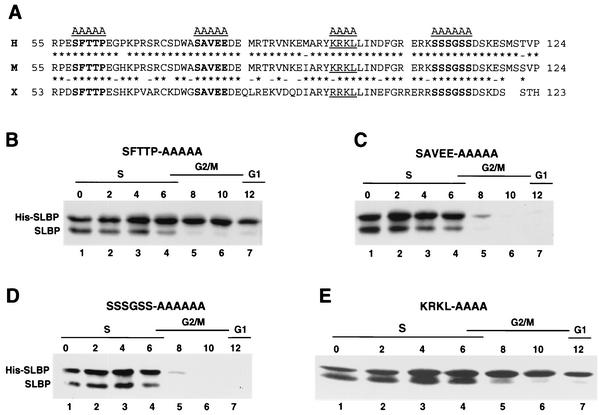
FIG. 2.
Amino acids 58 to 62 and 95 to 98 are required for SLBP degradation. (A) The sequences of the N-terminal regions of human (H), mouse (M), and Xenopus (X) SLBPs are shown (amino acids 55 to 124 in the human and mouse protein). The three conserved possible phosphorylation sites that were mutated are shown in bold type. The putative cyclin binding site (KRKL) is underlined. Conserved (asterisks) and similar (dashes) amino acids between two sequences are indicated. (B to E) Plasmids encoding the mutant His-tagged SLBPs (His-SLBPs) indicated above each panel were stably transfected into HeLa cells, and the cells were synchronized by a double-thymidine block. Protein extracts were made from the blocked cells (lanes 1) and from cells every 2 h after release from the block (lanes 2 to 7). Equal amounts of protein were resolved by SDS-polyacrylamide gel electrophoresis, proteins were transferred to nitrocellulose filters, and the endogenous and His-tagged SLBPs were detected by Western blotting with the anti-SLBP antibody. The time after release and the cell cycle stage is indicated above each lane.