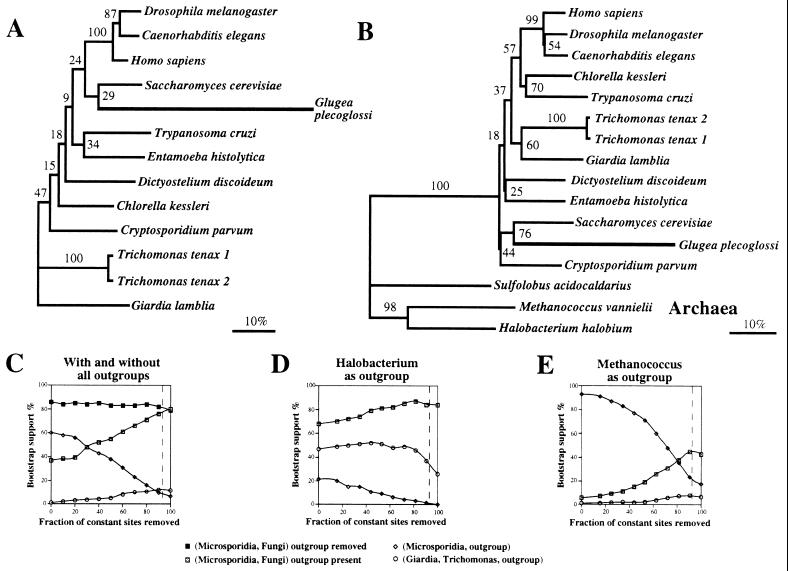
Figure 2.
Phylogenetic analyses of EF-2. (A) Protein ML tree with outgroups removed and FSR (72 sites). The scale bar represents 10% estimated sequence divergence under the JTT-F model. (B–E) Phylogenetic analyses of EF-2 DNA codon positions 1 + 2 using LogDet and investigating the influence of constant sites and choice of outgroup on bootstrap support. The DNA alignment was derived from a 542-aa alignment. (B) Bootstrap consensus tree topology for LogDet distances estimated from 812 variable sites (93% of constant sites removed as invariant) indicating support for M + F. The effect of incremental removal of observed constant sites on bootstrap support for different relationships in the presence or absence of all 3 outgroups (C), with Halobacterium halobium as outgroup (D), and with Methanococcus vannielii as outgroup (E). The dashed vertical lines represent the ML estimates of invariant sites: 93%, 92%, and 93% of constant sites for C, D, and E, respectively (see text for discussion).