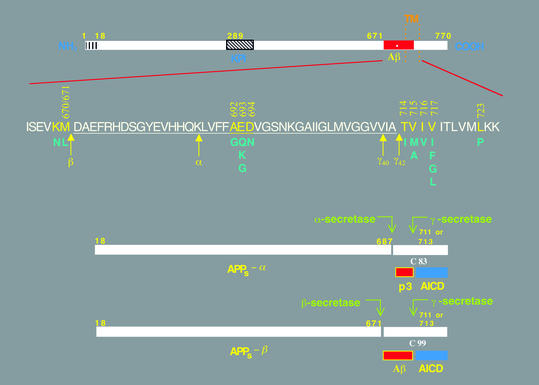
Figure 1.
Schematic diagrams of the APP and its principal metabolic derivatives. The first line depicts the largest of the known APP alternate splice forms, comprising 770 amino acids. Regions of interest are indicated at their correct relative positions. A 17-residue signal peptide occurs at the N-terminus (box with vertical lines). Two alternatively spliced exons of 56 and 19 amino acids are inserted at residue 289; the first contains a serine protease inhibitor domain of the Kunitz type (KPI). A single membrane-spanning domain (TM) at amino acids 700–723 is indicated by the vertical dotted lines. The Aβ fragment includes 28 residues just outside the membrane plus the first 12–14 residues of the TM domain. In the second line, the sequence within APP that contains the Aβ and TM regions is expanded. The underlined residues represent the Aβ1-42 peptide. The blue letters below the wild-type sequence indicate the currently known missense mutations identified in certain patients with AD and/or hereditary cerebral hemorrhage with amyloidosis. The three-digit numbers are codon numbers (βAPP770 isoform). In the third line, the arrow indicates the site (after residue 687) of a cleavage by α-secretase that enables secretion of the large, soluble ectodomain of APP (APPs - α) into the medium and retention of the 83-residue C-terminal fragment (C83) in the membrane. C83 can undergo cleavage by the protease called γ-secretase at residue 711 or residue 713 to release the p3 peptides. The fourth line depicts the alternative proteolytic cleavage after residue 671 by β-secretase that results in the secretion of the slightly truncated APPs β molecule and the retention of a 99-residue C-terminal fragment. C99 can also undergo cleavage by γ-secretase to release the Aβ peptides. Cleavage of both C83 and C99 by γ-secretase releases the AICD into the cytoplasm.