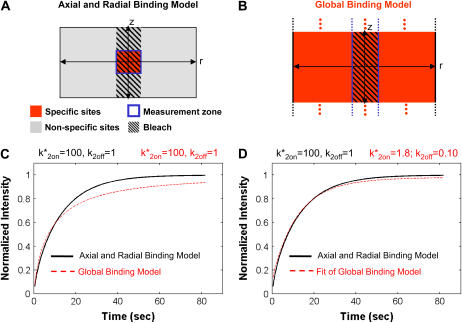
FIGURE 2.
Localized binding versus globally distributed binding. (A) The Axial and Radial Binding Model presumes a cylindrical (r,z) cluster of specific binding sites (MMTV promoters) 2 μm high and 2 μm in diameter, centered within a cylindrical nucleus 4.5-μm high and 15 μm in diameter filled with nonspecific binding sites and bleached with a cylindrical photobleach 2 μm in diameter (see Fig. 1 E). The FRAP recovery is calculated as the average fluorescent intensity within the specific binding zone, and corresponds to the measurement of intensity at the MMTV array from an optical section. (B) For the Global Binding Model specific binding sites are presumed to be distributed everywhere throughout the nucleus. In this model, variation in the axial dimension (z) is also ignored (dots indicate this symmetry in z). (C) Due to their different geometries, the two models generate different FRAP curves for the same binding parameters. (D) The Global Binding Model can be used to fit FRAP data generated by the Axial and Radial Binding Model, but the binding parameters predicted (red text) are quite different from the “true” values (black text). Thus the global binding model is a poor approximation to FRAPs at a cluster of binding sites. The FRAP curves in this and all subsequent figures have been normalized to their final recovery level. Units for k*2on and k2off in this and all subsequent figures are s−1.