FIGURE 1.
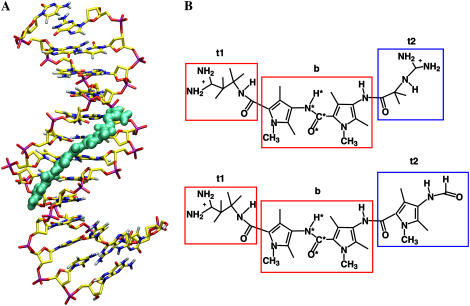
(A) Snapshot of the complex of netropsin with a d((CG)2A5(CG)2)·d((CG)2T5(CG)2) DNA duplex after ∼1 ns of MD simulation. (B) Chemical structure of netropsin (upper) and distamycin (lower) molecules. Rectangular boxes define the atom subgroups used to estimate configurational entropies: tail 1 (t1), body (b), and tail 2 (t2). Netropsin and distamycin molecules possess identical body (b) and tail 1 (t1) moieties (red rectangles), but differ in tail 2 (t2) (blue rectangles). The atoms of the central peptide bond (code 4) are labeled with a star. Reference codes for the entropy calculations are summarized in Table 1.