TABLE 3.
Internal configurational entropies of atom subgroups of netropsin and distamycin free in solution and bound to DNA, and correlations between their body and tails
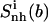 |
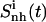 |
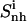 (t1) (t1) |
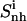 (t2) (t2) |
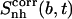 |
|
|---|---|---|---|---|---|
| Netropsin (free) | 384 (24) | 623 (41) | 363 (45) | 297 (42) | 157 |
| Netropsin (bound) | 333 (21) | 513 (34) | 290 (36) | 252 (36) | 113 |
| Δ (bound − free) | −51 (−3) | −110 (−7) | −73 (−9) | −45 (−6) | −44 |
| Distamycin (free) | 393 (24) | 651 (34) | 372 (47) | 328 (30) | 145 |
| Distamycin (bound) | 354 (22) | 568 (30) | 316 (40) | 296 (27) | 122 |
| Δ (bound − free) | −39 (−2) | −83 (−4) | −56 (−7) | −32 (−3) | −23 |
Corresponding changes upon binding are also reported (Δ). The body and tails of the ligands are represented in Fig. 1; Table 1 reports the reference codes. Only nonhydrogen atoms were used in the calculations. Least-squares superposition of structures was done using all nonhydrogen atoms. Per-atom entropies are given in parentheses. Correlation entropy 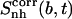 was calculated using Eq. 3. All values are in J K−1 mol−1.
was calculated using Eq. 3. All values are in J K−1 mol−1.
